Hi,
I have a question concerning loading covariance matrices.
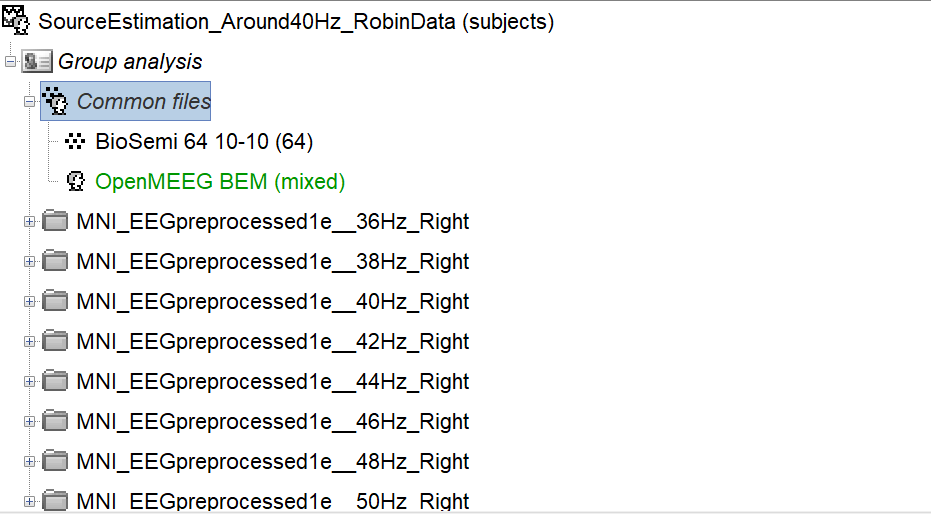
I have one subject called group analysis. I have some common files: the channel file and the head model and I have multiple sub-folders (one for each EEG data for each stimulation frequency that I used). See figure.
However, I would like to upload a different covariance matrix (that I already calculated in Matlab) for each sub-folder. To afterwards compute the sources for each sub-folder. Is this possible? Can I upload a different covariance matrix for each sub-folder?