Hello
This question might be more suited to SPM mailing list, let me know if that's the case. I have a T1 and T2 image of a subject that have been preprocessed using freesurfer. I imported the freesurfer folder in Brainstorm and segmented the T1 MRI using SPM12.
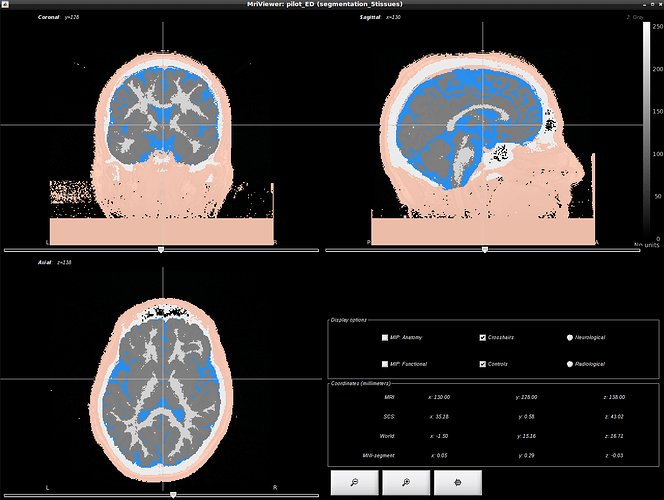
Unfortunately there seems to be some artifact in the background interpreted as skin. So i was wondering what would be the best way to clean this segmentation ? Would it be possible to use the head mask as a mask and to remove everything that is outside ?
Edouard
This is also my experience with the output of the SPM tissue segmentation. It requires some additional reconstruction.
Use CAT instead:
https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12
Or SimNIBS:
https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh?SimNIBS
Hello Francois,
Thanks for your reply. I will try Cat12. I just have a question related to that.
The description of the cortex in cat 12 is: cortex surface that was generated by CAT. The central surfaces are meshes half-way between the grey-white interface and the external pial surface.
it it equivalent of the mid surface generated by freesurfer ? (average between the pial surface and the gray matter-white matter interface)
Edouard
Description of the central surface on the CAT website:
We use a fully automated method that allows for measurement of cortical thickness and reconstructions of the central surface in one step. It uses a tissue segmentation to estimate the white matter (WM) distance, then projects the local maxima (which is equal to the cortical thickness) to other gray matter voxels by using a neighbor relationship described by the WM distance. This projection-based thickness (PBT) allows the handling of partial volume information, sulcal blurring, and sulcal asymmetries without explicit sulcus reconstruction (Dahnke et al. 2012).
Article describing the central surface estimation:
Thanks a lot François. I will have a look !