Hi,
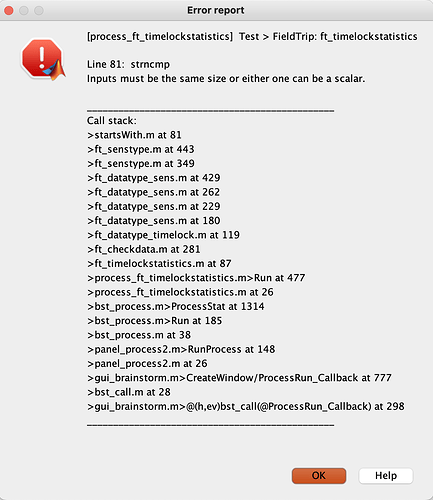
I received this comment from a reviewer on my paper: "In your EEG analyses. you used sample-by sample t-tests, corrected for multiple comparisons only at the electrode, but not at the sample level. This analysis procedure considerably inflates false discovery rate. If you want to stick to a sample-by sample analysis strategy, you should use non-parametric cluster permutation tests, which naturally control for false discovery rate. Alternatively, you should analyze EEG data in selected time windows at selected electrodes."
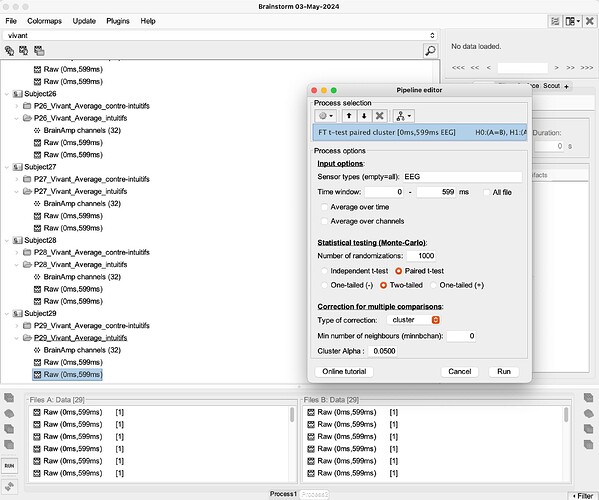
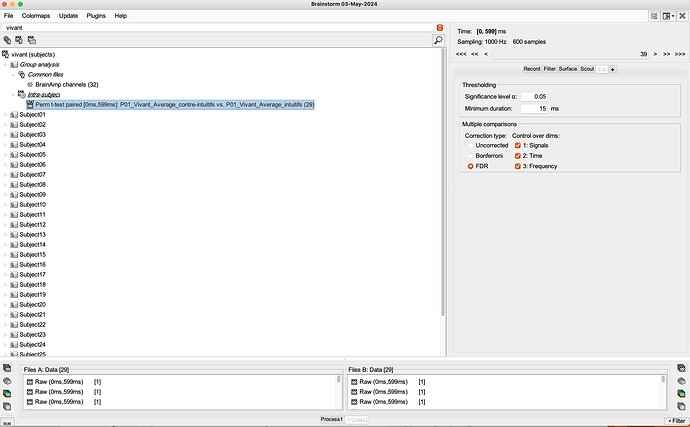
Here's what we've done: "Brainstorm software© [58] was thus used to statistically compare EEG signals from both intuitive and counterintuitive conditions, and make comparisons on all electrodes, at a p < .01 threshold with a Bonferroni correction for the number of electrodes. Only differences for specific time frames lasting at least 15 ms were considered at this step, in order to validate that the main differences in the signal were indeed localized only in the components of interest, and not on the whole signal. As a second step, top-down comparisons (paired sample t-tests) were run between conditions for the two components of interest. For exhaustivity purposes and since previous research found differences between conditions in various scalp regions [45], we ran comparisons for each main scalp region. We used the average of the electrodes located on a scalp region for both N2 and P3 components [45]."
I'm very new in this field and with Brainstorm, so I'm not sure if the reviewer is talking about our first or second step. Could somebody help me with that please? ![]()
Thank you!