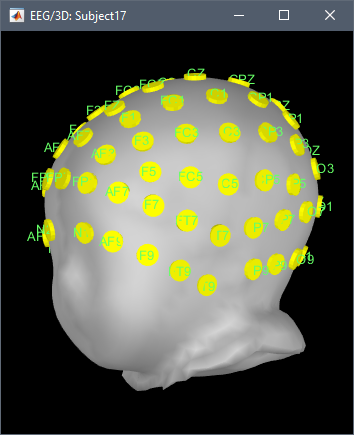
@Francois Channels and surfaces are already co-registered. It is relatively hard to have a small minimal program for demonstrating this, but here what it looks like (in Python):
Some imports:
from pathlib import Path
from nibabel.freesurfer.io import read_geometry
import pyrender
import mne
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import trimesh
And the main code:
# Getting the template
subject = "ANTS1-0Months3T"
subjects_dir = Path('/usr/local/freesurfer/subjects/')
mne.datasets.fetch_infant_template("1mo", subjects_dir=subjects_dir)
# Loading the "outer_skin" mesh
vertices, faces, metadata = read_geometry(subjects_dir / subject / "bem" / "outer_skin.surf", read_metadata=True)
head_mesh = trimesh.Trimesh(vertices, faces)
# Making a mesh of small spheres at the position of the electrodes
elec_pos_df = pd.read_csv(subjects_dir / subject / "montages" / "10-10_electrodes.tsv", sep="\t").set_index("name")
montage_meshes = []
for ch_name, (x, y, z) in elec_pos_df.iterrows():
montage_meshes.append(sphere_mesh([x, y, z], 2.5))
montage_mesh = trimesh.util.concatenate(montage_meshes)
# Showing the mesh
show_mesh(trimesh.util.concatenate([montage_mesh, head_mesh]), -1.5, 0, np.pi-0.5)
The result:
Full disclosure: To run, the code also need the definition of these functions (I just thought separating them would make the example more readable since they are not really related to the problem at hand, just to meshing and visualizing):
def show_mesh(mesh, angle_x=0, angle_y=0, angle_z=0, ax=None, resolution=(1200, 1200)):
mesh = mesh.copy()
Re = trimesh.transformations.euler_matrix(angle_x, angle_y, angle_z, 'rxyz')
mesh.apply_transform(Re)
scene = pyrender.Scene(ambient_light=[0.0, 0.0, 0.0],
bg_color=[1.0, 1.0, 1.0], )
scene.add(pyrender.Mesh.from_trimesh(mesh))
camera = pyrender.PerspectiveCamera(yfov=np.pi / 4.0, aspectRatio=1.0)
camera_pose = np.eye(4)
camera_pose[:3,3] = [0, 0, 240]
scene.add(camera, pose=camera_pose)
ligth_poses = [np.array([[-0. , -0.866, 0.5 , 0. ],
[ 1. , -0. , -0. , 0. ],
[ 0. , 0.5 , 0.866, 0. ],
[ 0. , 0. , 0. , 1. ]]),
np.array([[ 0.866, 0.433, -0.25 , 0. ],
[-0.5 , 0.75 , -0.433, 0. ],
[ 0. , 0.5 , 0.866, 0. ],
[ 0. , 0. , 0. , 1. ]]),
np.array([[-0.866, 0.433, -0.25 , 0. ],
[-0.5 , -0.75 , 0.433, 0. ],
[ 0. , 0.5 , 0.866, 0. ],
[ 0. , 0. , 0. , 1. ]])]
for pose in ligth_poses:
light = pyrender.DirectionalLight(color=[1.0, 1.0, 1.0],
intensity=1.0)
scene.add(light, pose=pose)
r = pyrender.OffscreenRenderer(*resolution)
color, depth = r.render(scene)
if ax is None:
fig, ax = plt.subplots(1, 1, figsize=(10, 10))
ax.axis('off')
ax.imshow(color)
def sphere_mesh(pos, radius, color=None):
mesh = trimesh.creation.icosphere(radius=radius, color=color)
mesh.vertices += pos
return mesh
Anyway, Brainstorm drove me nuts because the problem is so simple... everything is already co-registered. Just load it and use it. But somehow, I could not get Brainstorm to import these artifacts without breaking the co-registration.