I have several subjects, at each subject, I have 4 condition (4 different sensor montage)
I want to compute the gain matrix for all of them using openmeeg at one click, ist it possible?
Interactively: Right-click on the subject instead of the folders.
Recommended approach: Write a script.
https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting
This won't work if you have warnings.
Generate a script instead.
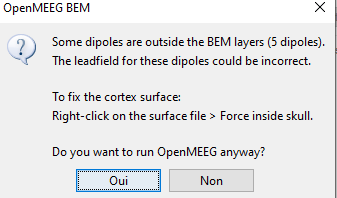
I am going to each subject one by one and make force the cortex inside...but even with script I shoud do this before?
generating a script seems to be difficult for me, I tried but I am not succeed even to compute the leadfield for one case
please help me just in one subject, because I always have this error:
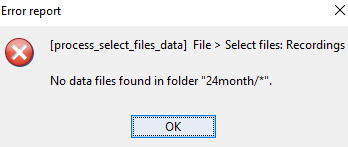
steps that I did:
- i click run in process 1 where it is empty,
- file->select file recording
- source-> compute head model
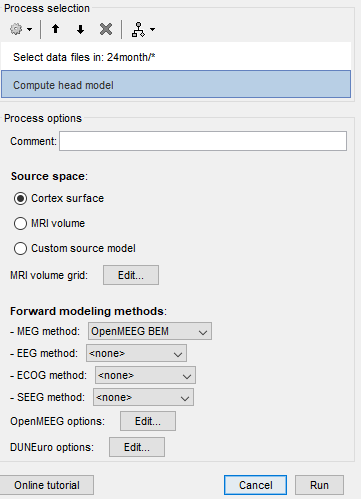
- run
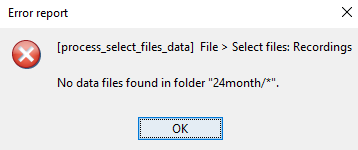
Because you don't have any data files in there, indeed.
The pipeline editor and Brainstorm batch system can only have in input and output files containing signals (recordings, sources, time-frequency, scouts signals...). To be called from a script, the "Compute head model" process needs that you import of simulate some signals in there.
I guess what you are trying to do here is some simulation work on forward models only.
This very specific case can't be handled by the standard Brainstorm scripting system. If you don't have any recordings in the folders in which you want to compute forward models, you need either to do it from the interface, or to call the lower-level functions manually. Two options, depending on the level of details you want to provide:
-
panel_headmodel('ComputeHeadModel', iStudies, sMethod): Read the help at line 284 inpanel_headmodel.m -
bst_heamodeler.m: Put a breakpoint at line 673 panel_headmodel.m, observe what is in the input structure and reproduce it in your script.
Not that these are advanced undocumented features of Brainstorm: we can't guarantee backwards compatibility for these calls in the future, and we can't provide step-by-step help for using them. You need to be very confident with your Matlab programming skills to try either of these solutions.
Having vertices outside the inner skull surface is a problem for OpenMEEG.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem#Warning:_Dipoles_outside
You can either force the vertices of the cortex surface to be inside the inner skull: see function tess_force_envelope.m
Or you can recompute another set of BEM surfaces with Brainstorm, using a higher number of vertices.
If you are using @christianoreilly 's templates, I guess the BEM surfaces were created using a different algorithm, probably MNE's mne_watershed. @christianoreilly Is this correct?
Recomputing the BEM surfaces in a different way (as in the beginning of the BEM tutorial) might solve your problems.
It seems that just one subject have the problem, I solved it by force inside skull and now I am computing the gain matrix for all of them
does this mean that I cannot used openmeeg?
It means that you need to be careful about this issue:
https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem#Warning:_Dipoles_outside
@Francois Not the watershed algorithm because I already had the voxels labeled (scalp, skull, ...) so the surfaces were obtained by marching cubes + Laplacian smoothing + mesh decimation + corrections for topological issues like holes and such...
Not the watershed algorithm because I already had the voxels labeled (scalp, skull, ...) so the surfaces were obtained by marching cubes + Laplacian smoothing + mesh decimation + corrections for topological issues like holes and such
For some subjects, the cortex and inner skull surfaces seem to be intersecting, which might cause problems running OpenMEEG.
Have you computed forward models with OpenMEEG with all the age ranges?
There should be no intersection. Are you sure OpenMEEG does not check for a minimal distance between these surfaces? I did not use OpenMEEG. I used MNE-Python on these models. It also crashes when surfaces are intersecting. All the models run through my code which checks for intersecting surfaces and "push" the outer surface out of the inner one and then they have been check with MNE-Python and they were working fine, not intersecting. I did not set a minimum distance (like >1 mm) between surfaces so if Open MEEG apply such criteria, it might complain... one model in particular is causing problem for you?
@abdallahqusaibe Can you please tell us how got this warning?
Please tell us which template you used, and how step by step.
Please include screen captures showing the anatomy files in the database explorer, all the option windows for starting OpenMEEG, and a 3D view showing the inner skull + the cortex + the electrodes.
Thanks
@Francois Thinking of it, I should point out that what I said applies to the last release of the templates and I am not sure if these have been integrated in Brainstorm or if what is available in Brainstorm is an older version. At what date did you package the templates currently available in BS? I tried to integrate the corrected templates into BS around Nov 25th 2020 and did not manage to get it anywhere with the import script (I could not make sense of the coregistration information BS was requiring). So, bottom line, if the current version of the templates has been packaged before mid-November or so, it is probably an older version of the templates and I cannot say for sure if these may have intersection issues.
@christianoreilly
The packages downloaded from the Brainstorm are from July 8th, 2020.
We worked on it in November 2020, but stopped without pushing any updates:
Yes... I looked at it for 1-2 days and could not make sense of how Brainstorm expected the data to be provided so that it can import it consistently. I can try to have a look at it again when I will have a bit of time. In the meantime, if you want to have a look at the import script ( https://github.com/christian-oreilly/infant_template_paper/blob/master/import_to_brainstorm/import_template_to_brainstorm.m ) and check if you can figure out what is going wrong, your help would be appreciated. I just committed and pushed this file, as it was last time I worked on this... so it is not a "clean" commit but it should give you the current state of the code I was trying to fix.
If I remember well, the different data could not be set on a correct scale... I started to write you a message back then and I was trying to make sure everything made sense before sending it and I guess I just did not manage to reach a level of understanding of what was going on that was satisfying for me to complete the message... Here was the (unfinished) message in case it can help explain the issue (that was in continuation of a discussion at https://github.com/christian-oreilly/infant_template_paper/issues/1# :
I will phrase it in my own words (which might end up being just what you are trying to tell me, but please bear with me) so that I can be sure that I understand: The only way I see that it can work is with sMri.SCS.T in mm (as mentioned by the comment "This hack was added becaue cs_convert() is intended to work on sMri structures, in which NAS/LPA/RPA/T fields are in millimeters") and consequently sMri.SCS.T./1000 is in m. The transform is therefore applied to coordinates that need to be in m. So, in
ChannelMat.SCS.NAS = cs_convert(ChannelMat, 'mri', 'scs', ChannelMat.SCS.NAS ./ 1000) .* 1000;
of channel_detect_type, ChannelMat.SCS.NAS needs to be in mm so that ChannelMat.SCS.NAS ./ 1000 is m and is in compatible unit with sMri.SCS.T./1000. So... the last place that seems not to work for me is when you way "ChannelMat.SCS.NAS / .LPA / .RPA are in SCS coordinates (meters)".
Again, as per the comment "NAS/LPA/RPA/T fields are in millimeters, while in ChannelMat they are in meters", if ChannelMat.SCS.NAS is in meters, then ChannelMat.SCS.NAS ./ 1000 is in km, whereas sMri.SCS.T being in mm, then sMri.SCS.T./1000 is in m and these two things are off by a factor 1000.
That part works if we do something like (with sFid as used for import_anatomy_fs(iSubject, FsDir, 15000, 0, sFid)):
ChannelMat.SCS.LPA = sFid.LPA;
ChannelMat.SCS.NAS = sFid.NAS;
ChannelMat.SCS.RPA = sFid.RPA;
which fits with your previous statement "When you import the MRI (before importing the surfaces), the NAS/LPA/RPA must be set to points that are matching EXACTLY the positions of the NAS/LPA/RPA defined in the .tsv positions file."... but in that case, ChannelMat.SCS.NAS is indeed in mm, not in m. With that, I obtain channels and surfaces that are on the same scale (obviously the transformation is still wrong though, for whatever reason...):
But if instead I use
ChannelMat.SCS.LPA = sFid.LPA / 1000.0;
ChannelMat.SCS.NAS = sFid.NAS / 1000.0;
ChannelMat.SCS.RPA = sFid.RPA / 1000.0;
so that these are in m, then I get [... and I am afraid I stopped writing there...]
This is terribly confusing, I don't know where we are anymore.
Could we start again from a simple starting point?
- Upload somewhere a clean anatomy for only one of you age templates (or tell me where to download it from), in whatever file formats you are primarily using to distribute it. Something stable, reliable, published.
- Describe precisely what needs to be done on it in Matlab/Brainstorm and exactly how you'd call you conversion code (
import_template_to_brainstorm.m?) - Describe what you expect to obtain and what you obtain instead
Subsequent steps:
- I'll take it from there and debug it to make it work on one single age range (and probably ask you more details about the contents of the template)
- I'll send you back the modified import function
- You'll re-run it on all the age ranges
- You'll generate the Brainstorm anatomy templates (right-click on subject > Use template > Create new template)
- You'll upload all the updated .zip file somewhere
- I'll upload them to the Brainstorm webserver and update the online documentation and forum posts.
Does that sound like a reasonable approach?
Hi @Francois. Sorry for the late reply.
Upload somewhere a clean anatomy for only one of you age templates (or tell me where to download it from), in whatever file formats you are primarily using to distribute it. Something stable, reliable, published.
The binary release are available for download here: Release Template binary release · christian-oreilly/infant_template_paper · GitHub
They are working fine in MNE. Importing them in Brainstorm is problematic as the co-registration does not work for some reason and I could not make sense of what Brainstorm was expecting from BS code.
Describe precisely what needs to be done on it in Matlab/Brainstorm and exactly how you'd call you conversion code (
import_template_to_brainstorm.m?)
Whatever needs to be done for these to be used as templates in Brainstorm. I was simply running import_template_to_brainstorm.m (it is not a function to be called, just a script to be run once to prepare the templates) and the expected outputs are the .zip files that you mention in the second-to-last point for "subsequent steps".
Describe what you expect to obtain and what you obtain instead
It's been too long for me to remember the details, but if I remember well, the surfaces were just not co-registered with the surface as they should.
You'll generate the Brainstorm anatomy templates (right-click on subject > Use template > Create new template)
The idea is for that to be part of the script. The idea is to avoid any point-and-click kind of operation and rather have the process scripted so that it can be part au the release pipeline.
I've written a new script for preparing these anatomy templates in Brainstorm:
bst_create_templates.m (4.5 KB)
I think there are a few minor issues with the BEM surfaces distributed in these atlases:
- The outerskin surface is a bit too large with respect with the actual skin (I guess this is an MNE issue)
- The innerskull apparently suffers from some issue in the brain extraction (see the little horns below).
I have another major problem with the electrodes positions.
I would like to read the _electrodes.tsv you provide in the montage folder, but they don't seem to be in the MRI referential... What are the coordinates in the .tsv files referring to?
If it were BIDS-compatible, it would need to be in the "scanner-based" coordinates of the reference MRI mri / T1.mgz.
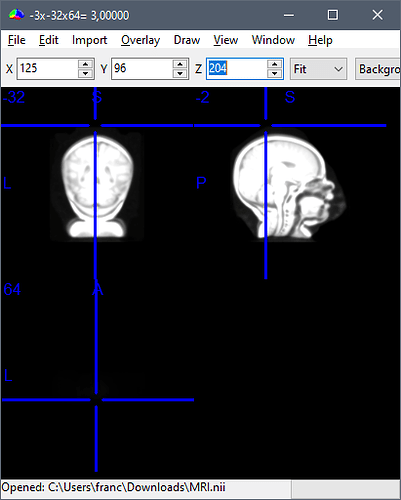
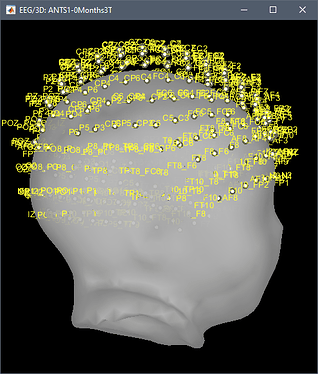
Example for CZ electrode in ANTS1-0Months3T / montages / 10-5_electrodes.tsv:
CZ -3.0558934522120014 -32.064090002355826 64.51582182251714
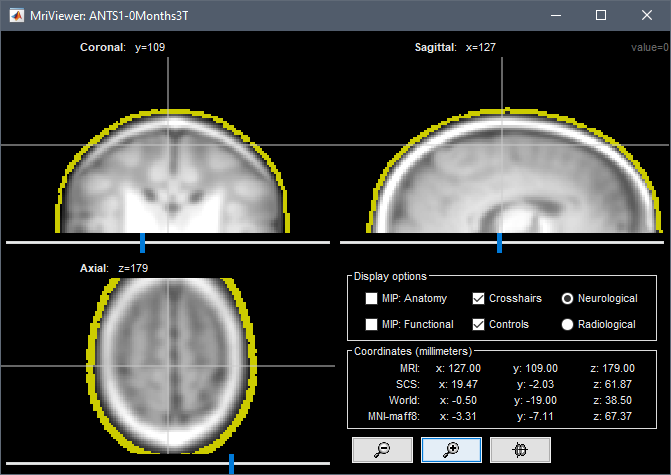
But if we input these coordinates in the MRI scanner-based coordinates of T1.mgz, we get this location:
This results in an incorrect registration in Brainstorm: