Hello Francois,
I have another question. I am collection Brainvision data that I prepare in EEGlab and then import into Brainstorm.
I have a strange issue of an event shift of 1 sample.
The data sample with the event that should happen at the 0.000 time mark of the epoch I am importing appears at -0.001 instead.
To me everything still seems in order at the time I am exporting the data from EEGlab. The data sample in EEGlab is at the 0.000 mark.
Do you have any idea what might be the issue here?
What information does brainstorm take from the set/fdt files to line up events?
Thank you!
Christian
Can you please share an example EEGLAB file?
Zip it, upload it somewhere, and post the download link here.
Please include a screen capture of what it looks like in EEGLAB (the timing t=0).
Thanks
Thank you Francois for your response.
Here is an example where it even seems to be more than 1 sample.
First the way the data looks in EEGlab when I save the dataset.
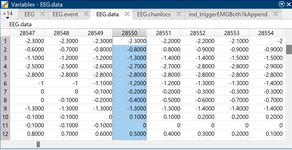
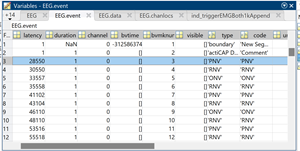
The latency in EEG lab identifies the event as the number of samples. So I would expect the highlighted sample to appear at the 0.000 position.
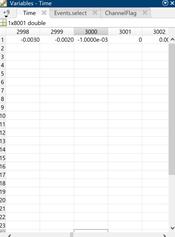
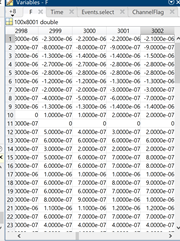
Now here is what the data looks in brainstorm after input (opening the epoch file in Matlab). The event I selected is the first PNV event. First the Time variable which makes me believe I would see the 0.000 sample at index 3001, however looking at the F variable it seems to me in this case the variable is in 2999 2 samples earlier.
Here are the set/fdt files used attached. Even as a zip file it was too big for this forum, let me know if this share link does not work for you.
https://musc.box.com/s/c4f0x613nuk40xza19s5zmo86qnpvywh
Thank you so much for your assistance!
Christian
Can you please upload the file to a service that does not require a paid account to download the file? (dropbox, google drive, ...)
I also find it quite difficult to follow the description of the problem from these screen captures. Hopefully it will make more sense with the data in hand.
Can you please add screen captures of:
- what looks wrong in the interfaces of both EEGLAB and Brainstorm. What part of the interface do you look at to conclude that there is something wrong?
- all the steps to import this file into Brainstorm (menus, option windows...)
Thanks
Here is a google drive link: example.zip - Google Drive
When comparing the information in the interface, the 1-2 sample shift is not visible. Even with the shift, the data successfully imports and epochs into brainstorm. However, when I compare the data points in EEGlab & Brainstorm at the event (T=0.000) to confirm consistency, the data do not match.
The first two screen shots in the previous message show me grabbing the 0.000 data point from EEGlab (they store events in EEG.latency as datapoints and EEG data in EEG.data). Based on the EEG.latency variable, the 0.000 sample of the first event should be in column 28552. The other two screen shots show me grabbing the 0.000 data point from the epoch of the first event in Brainstorm (data.mat file of the epoch using the Time and F variables). Based on the Time variable, the 0.000 datapoint should be in column 30001 of the F variable, but these data do not match with EEG.data in column 28552 (there are different values in the first 12 channels visible in the screenshot). You can also see that the sample to result in a successful match at -0.002 seconds (ID 2999). Therefore, this is a shift of two samples between the EEG.data variable and the F variable.
Looking more at the data from multiple trials it seems to me that the first 1-2 data samples do not show up in brainstorm but it uses the same events therefore I get a shift.
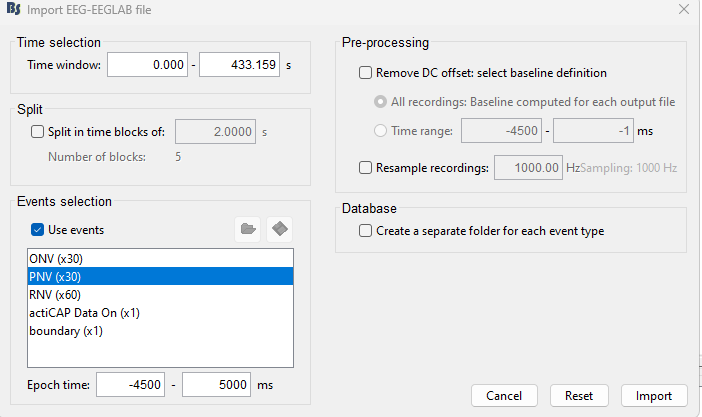
Here are screenshots of the full import process.
After clicking on the "Import MEG/EEG" on the session folder I get the following prompt:
Then I epoch based on events
This imports the data.
I hope this explanation and link was better than the last one and works.
Christian
I think there might be some confusion in what you are considering here.
What you describe mixes two processing steps:
-
The reading of a EEGLAB .set file into Brainstorm as a CONTINUOUS file: the event markers should point at the same sample both in Brainstorm and EEGLAB. Note that this does not mean necessarily the same sample index, or event the same time value, as time references are conventions and can be different across applications (e.g. first sample index can be considered as being labelled 0 or 1, conversion between time and sample can be different...)
=> If you suspect a problem at this level, you should explore only the TIME values (not the sample indices), in CONTINUOUS files (using the menu Review raw file instead of Import MEG/EEG), as displayed in the INTERFACE of both EEGLAB and Brainstorm.
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Review_vs_Import
-
The epoching process in Brainstorm, i.e. extracting a short time segment of data from a continuous file around an event of interest (https://neuroimage.usc.edu/brainstorm/Tutorials/Epoching). This is what you do by using the menu Import MEG/EEG. The number of samples that is captured before and after and what is considered as sample #1 or time t=0s are conventions that might different between Brainstorm and EEGLAB. In Brainstorm, the sample that is pointed by the marker used for the epoching will be considered as t=0s in the epoched file, no matter how many samples are actually imported before or after. Trying to understand timing based on sample indices in epoched data can only lead to confusion.
=> It is unlikely that there is a problem in the way Brainstorm does its epoching.
I don't really know to help you here, as if you can't show any mismatch in the interface, the problem is maybe in your interpretation of the sample indices. To explore in an appropriate way what you suspect to be a bug, I suggest you start by observing what are the values at one occurrence of one of the events, both in Brainstorm and EEGLAB and using the interface (and not relying on your possibly erroneous interpretation of the underlying data structures). If you find a mismatch here, then there could be 1-sample error in the interpretation of the EEGLAB events in Brainstorm (2 samples would be a lot more surprising).
Make sure you don't have any online filters, ICA selection or montages applied.
Then in you doubt about the second step (epoching): with the Brainstorm interface only, compare a) the EEG values at the marker in the continuous file with b) the EEG values at t=0 in the epoched file.
Make sure you don't apply any computation to the epochs that are imported (like in the screen capture of your last post: no ICA, no SSP, no baseline correction, no resampling).
Please get to us after performing these checks.
This actually helps more than you think Francois! I think I know what to do now!
Thank you so much and sorry for my limited understanding at first and difficult explanation. Thank you for all your time and patience!