I am getting the following error when I run process_pac from the GUI (i.e., Frequency > Phase amplitude coupling)...
** Error: [process_pac] Frequency > Phase-amplitude coupling
** Line 362: Array dimensions must match for binary array op.
**
** Call stack:
** >process_pac.m>Run at 362
** >process_pac.m at 29
** >bst_process.m>Run at 230
** >bst_process.m at 37
** >panel_process1.m>RunProcess at 124
** >panel_process1.m at 26
** >gui_brainstorm.m>CreateWindow/ProcessRun_Callback at 781
** >bst_call.m at 28
** >gui_brainstorm.m>@(h,ev)bst_call(@ProcessRun_Callback) at 305
**
**
I've been able to determine that the error is occurring when line 362 of process_pac.m tries to execute the following line:
DirectPAC_avg = DirectPAC_avg + DirectPAC ./ length(sInputA);
The issue seems to be that:
size(DirectPAC_avg) = 64 1 6 17
size(DirectPAC) = 63 1 6 17
length(sInputA) = 77
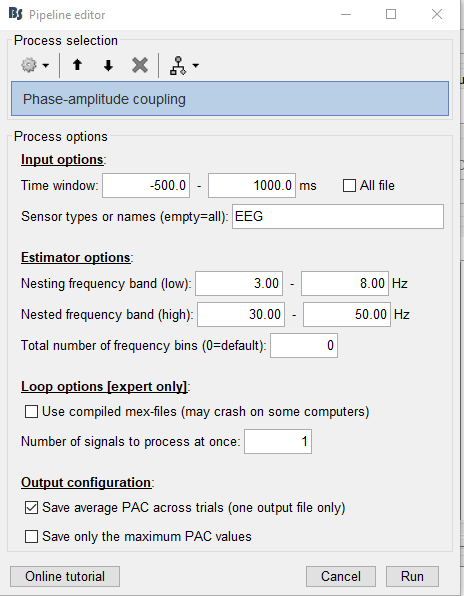
The parameters I'm using are below:
Can anyone help me figure out the issue?
It looks like not all your files have the same number of channels (some 63, others 64).
To check that this is the error: try computing a simple average, it should fail as well.
Then identify the files with less channels, and fix the problem (possibly with the process "Standardize > Uniform list of channels")
Francois,
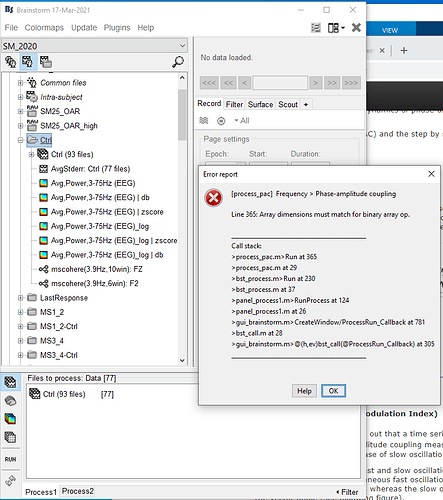
Thanks for the quick reply. Strangely, I have been able to average files, compute average wavelet transforms and even average coherence all with the same set of files! It is only process_pac.m that is giving me trouble. See image below. I'm trying to investigate whether "bad" channels could be the issue.
Francois, It does seem to be the case that process_pac.m is removing bad channels on a trial by trial basis. My first trial has one channel marked as bad so N=64, but the second trial has two marked as bad, N=63, which lines up with the error. I would like to ignore the bad channels in the average PAC. Can you suggest a workaround? Many Thanks!
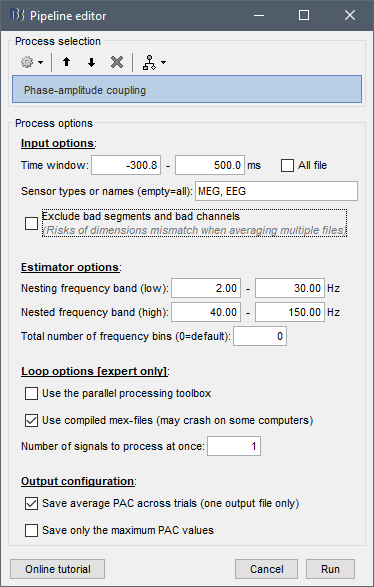
Indeed, by default the reading functions used in process_pac.m exclude both the bad segments and the bad channels, which is useful in some cases but annoying in others.
Therefore, I added a checkbox in the process to make it an option.
If you uncheck it, it won't crash anymore, but you have to keep in mind that you have some channels for which the estimation is maybe inaccurate.
Update Brainstorm to get this new option.
Alternatively, you can mark the channel as bad in all the input files (select them all at once in the database explorer > Good/bad channels > Mark some channels as bad). This way you be sure the bad channels are exclude from the computation.
Thanks Francois, this is exactly what I was hoping for. That said, this option is already available for the Coherence functions and I wonder if there is any reason to think that PAC estimates would be more inaccurate with bad channels included as compared with Coherence. My thinking is that excluding bad channels makes sense for ERP and even ERSP, but Coherence and PAC are usually estimated for specific frequency ranges that do not often overlap with the frequences producing major artifacts. Thoughts?
Different processes written by different people at different times might have different lists of inputs for no particular reasons... 
It really depends what "bad" is. If you are processing SEEG and you have half of your electrodes that were not plugged, you don't want to spend time computing complicated measures on empty signals.
Also, this concerns bad SEGMENTS. You may have periods where the subject is moving that you'd like to remove.
The configurations are infinite, we're trying to provide tools that can satisfy everybody, up to you to find what is the correct combination for your data.
Thanks Francois,
I completely understand. I've used Neuroscan, Analyzer, and EGI's software and was a long time user of EEGlab. I have to say that Brainstorm has quickly become my favorite.
Francois,
Thanks again for adding the option in process_pac. It doesn't seem that an update is available yet. Will one be released soon?
Paul
The commits pushed to the Brainstorm repository are available as updates at most 24hr after:
https://github.com/brainstorm-tools/brainstorm3/commits/master
The menu "Update > Update Brainstorm", will give you access to this fix.
Unless you are using the compiled version of Brainstorm?
In that case, you will need to be more patient, I will not push these update immediately.
To fix this issue, you have the option to mark the bad channels as good or as bad in all the files.