Hi people! I’m a beginner in using Brainstorm. To plot a .mat neuromag file which contains positions (x,y,z coordinates of sensors i.e. a 318 x 3 dimensional vector) I downloaded the sample neuromag file from brainstorm website. But the downloaded file has 3 x 4 dimensions for position for each sensor.
I’m hence having trouble to map my 1 x 3 dimensions to 3 x 4 dimensions for each sensor. Can anybody help me? Thank you!
Hello
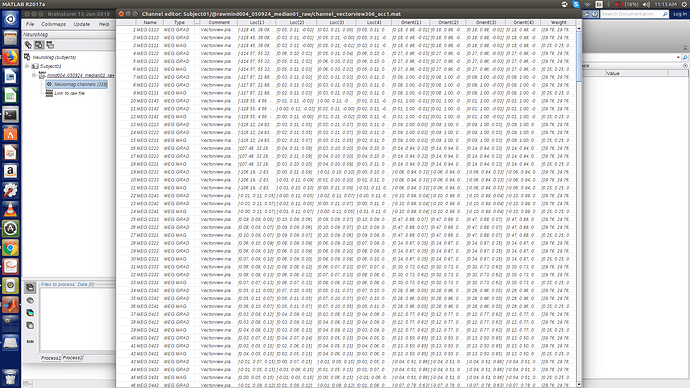
Brainstorm uses 4 integration points for each MEG sensor (corresponding to the 4 corners of the Elekta magnetometer or gradiometer).
However, if you use only one point per each location, all the forward/inverse modeling functions should still work. The easier for you is probably to fill the .Loc structure in the channel file with only one 3D position (make sure you keep the correct matrix orientation: [3x1]).
You should adjust accordingly the Orient field (the normal to the sensor plane) and the Weight field (you can set it to [1], or leave it empty [], I think both options should work).
Francois
Hi Francois, thank you so much for replying!
If I fill up my 3D position in one of the .Loc structure, what do I fill up in the other three?
Also, to change the .Loc, Orient and Weight attributes, will I have to manually update them for all 306 sensors in the table?
You need the orientation of the sensors if you want to be able to process them correctly… You don’t have this information? If you set the Orient field to a random value (eg. [1;0;0]), you’d be able to plot 2D/3D topographies, but not to do any source reconstruction.
Pay attention to the type of the sensors: MEG MAG (sensors ending with “1”) or MEG GRAD (sensors “2” and “3”).
I have the Orientation of the sensors also in 3x1 format, and not in 3x4 format. So what do I fill up in the other 3x3 columns?
Do the same as for Loc: use only one orientation [3x1].
Would I have to manually update all the rows and columns? or can I update them through some lines of code in editor?
Also, can I highlight some particular sensors through MATLAB editor? If yes, what code should I write in such cases?
If this is my channel editor window, if I update my current 3x1 position matrix in Loc(1), what data do I fill up in Loc(2), Loc(3) and Loc(4)?
You need to do this from a Matlab script, you won’t be able to this from the Brainstorm interface.
Load the file with the load() command, modify the structure, save it back with the save(…, ‘-struct’, ‘variable_name’) function.
The structure of the channel files is described here:
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Edit_the_channel_file
If you are not familiar enough to do this with Matlab, why don’t you just load the original .fif files?
That would be by far the easiest solution…