Hello,
We find difficulties opening .mat file of an MEG recording in Brainstom, it's open only as a EEG file, but didn't give the true signal in fTesla
when we open it as MEG It doesn't work,
we are thinking of solutions : for exemple to copy the matrix element from the .mat file and paste it in a .fif m file , is this possible ? how we can open an .fif file ?
thank you in advance
First of all, a .mat file is not a file format for MEG data, it is a generic contained with an unrestricted list of variables. There is no way for any program to guess what is it.
If you file is a SPM or EEGLAB file structure, read it with the correct file format selected instead of the the generic ".mat" entry.
If your .mat file contains only the sensor signals saved as a 2D matrix, it should not be used to import MEG recordings into the Brainstorm database. There are lots of extra information that are needed to process this data correctly in the original files (sensors positions, types, properties, calibration of the references, etc). There is no easy solution for doing is in Brainstorm, because it should not be done this way. I recommend you import the original MEG recordings in Brainstorm instead.
If you still want to do this, you need to do everything manually, and probably get some proper training with Brainstorm data structures and scripting before: https://neuroimage.usc.edu/brainstorm/Tutorials/Scripting
- You would import you data as EEG
- You need to edit the channel file and change the types of the sensors to MEG
- You can't enter the positions/orientations of the sensors and therefore won't be able to perform any source localization, or to display 2D/3D topo plots
- The only thing you'd be able to do is to display the signals, and run some processes on them (e.g. averaging them). You can already do this in Matlab, you don't need Brainstorm for that.
we are thinking of solutions : for exemple to copy the matrix element from the .mat file and paste it in a .fif m file , is this possible ? how we can open an .fif file ?
FIF is a complicated file format. I don't recommend you try in this direction if you are not an expert in this field.
Thank you so much Francois for your help.
Your answer was very complete,
We have raw data of MEG acquisitions with OPM He4 of 5 sensors in the axes X,Y and Z ( 15 in total ) , we use 2 files (.mat) for the matrix and (.json) and import them in python and then save them into a .fif file.
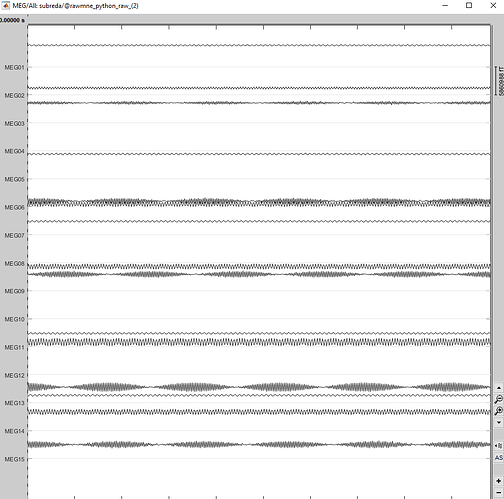
then we use this same file to plot the signals in Brainstorm and when we click " link to raw files ' we get 15 periodic signals, when we normally get a aleatory signal with the SQUIDS signal.
do you have any Idea why there is this problem ?
Thank you so much.
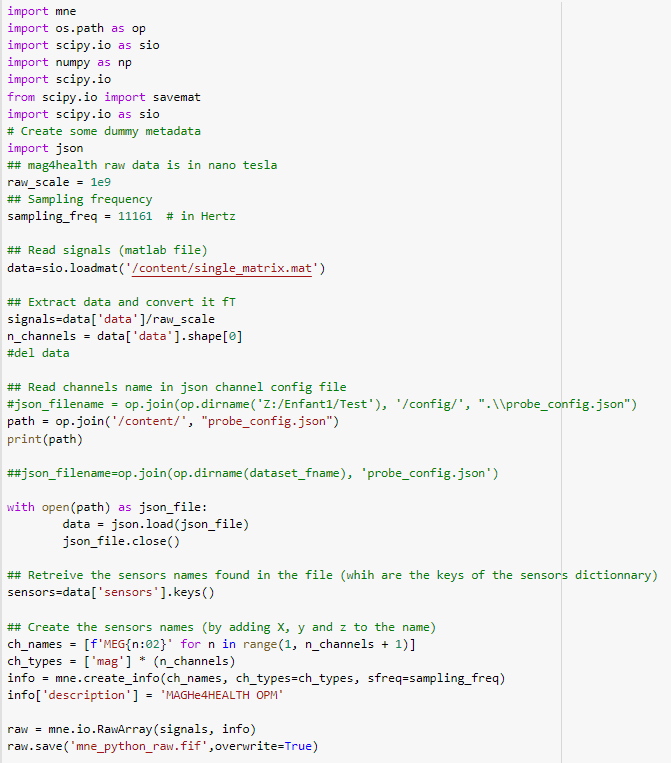
the program in Python :
the results of the link to the raw file .fif
Hi @redaoumiel,
It seems the issue is in the data in the .mat file.
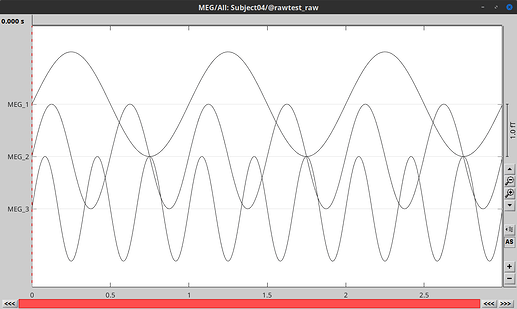
I created a .fif file with MNE-Python and linked into Brainstorm without any trouble
Code: https://gist.github.com/rcassani/abb2eaa3cabb135eb0f15a8aab0ff6a8
I can see, Thank you Raymundo.