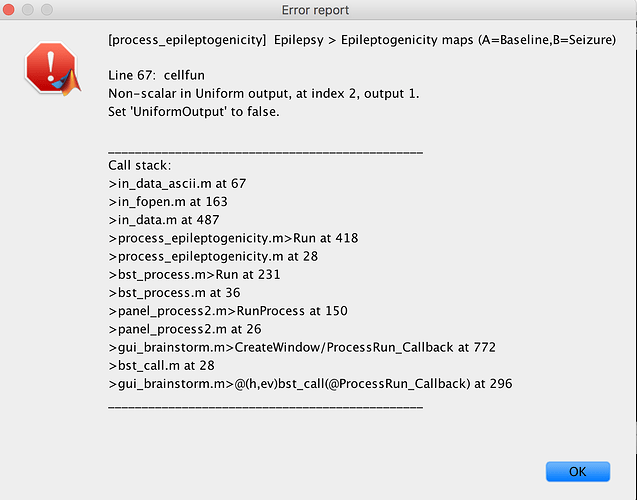
Dear Francois,
I get the following error while performing epileptogenicity maps on brainstorm with SEEG data.
I have followed the tutorials thoroughly and I haven't got any problem so far (could it be related to some sort of conflict with SPM?)
Thank you in advance for your support,
Best Regards
Sorry for the response delay.
If you have are using other toolboxes in Matlab, there might be conflicts between some functions:
- First make sure you are using the latest versions of Brainstorm (menu Update > Update Brainstorm) and SPM12 (execute
spm update)
- Edit your Matlab path, and remove all the folders that are not part of the Matlab installation
- Close Matlab and restart it
- Go to the brainstorm3 folder, do not add it in your path, and run brainstorm.m
- In the Brainstorm preferences, check the path to the SPM toolbox (do not add it to your Matlab path either, this is all done automatically)
- Try again running the epilptogenicity computation.
It might very well be that the error you have is not due to path issues. It could be an error manipulation on your end, or a bug in the software.
- If you haven't followed the Brainstorm introduction tutorials (section "Get started"), start by following them all at least until #19, in order to get familiar with the software
- If you are using your own SEEG recordings and have not followed the tutorial with the example dataset: start again with the example dataset provided to get familiar with the software before trying to process your own data.
- If you are following the epileptogenicity tutorial using the example dataset provided: delete the protocol, start over from the beginning, and make sure at each step you obtain exactly what it shown in the tutorials.
If after doing all this you still obtain exactly the same error: I would need an example dataset in order to reproduce and investigate the error:
- Right-click on the subject > File > Duplicate subject
- Delete all the files that are not needed to reproduce the behavior (both in the anatomy and the functional folder)
- Make sure you can still reproduce the error with this minimal dataset
- Right-click on the subject > File > Export subject
- Upload the zip file somewhere (dropbox, google drive...) and share the download link here (or send it to me via private message if this is sensitive medical data - you can also also deface the volumes before sharing them: right-click on the MRI > Deface volume)
- Describe exactly how you call the process in order to get this error (include screen captures of the process options)