Dear brainstorm experts,
Hellow,
I have build both "constrained" and "mixed" forward models for my subjects, but when visualizing the results of source localization, it seems to be a problem with the "mixed" model for one subject; only one hemisphere gets activated. With the simple constrained model, both hemispheres get activated as expected.
Would you please help me with this?
Can you please post screen captures illustrating your problem?
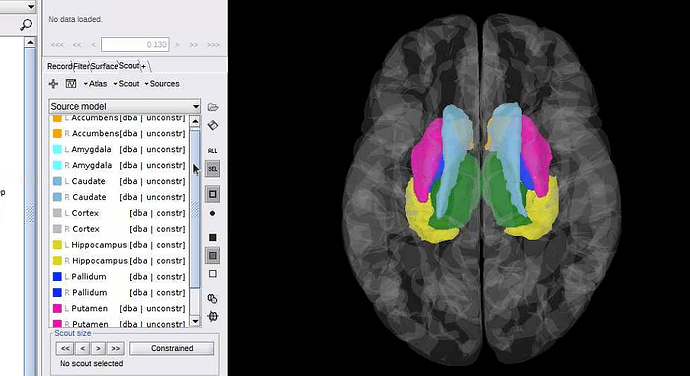
- The contents of your atlas "Source model" in the Brainstorm window, and their view in 3D (as this tutorial: https://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas#Select_deep_structures)
- Right-click on the forward model > View lead field vectors > Follow the instructions to djust the display in order to see whether the arrows look similar in the left and right hemispheres => Screen capture
- The source results with the Amplitude slider set to 0%
- The MEG or EEG recordings time series, with the red cursor at the same time as the source maps
- All this both for the constrained and mixed models.
Note that we don't really recommend using this mixed models - see the warnings at the beginning of the tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas
Dear Francois,
thanks a lot.
I am attaching the requested images, but of course the lead field vectors are not shown properly, although I tried much, sorry for that.
I had seen the warnings with the mixed model, but there are two observations based on which I have still preferred using it:
1- when projecting single subject results to the common space, the brainstorm shows a high overlap and nice match between the final projected subcortical structure and the assoicated one in the standard brain for most of the subjects.
2- It seems that I get almost good results in the group level analysis since they are consistent with results of fmri (taken simultaneously).
Is there other serious theoretical or technical problems with this "mixed" method that I shall revise using it?
Best regards,
EEG_3D_subj_stims_final_Avg_20_files_MN_EEG_Constr_2018.tif (439.8 KB)
EEG_3D_subj_stims_final_Avg_20_files_MN_EEG_Mixed_2018.tif (426.3 KB)
Leadfield__users_EEG_brainstorm_db_Porotocol01_data_subj_@default_study_headmodel_surf_openmeeg.mat.tif (186.4 KB)
Leadfield_users_EEG_brainstorm_db_Porotocol01_data_subj_@default_study_headmodel_mix_openmeeg.mat.tif (220.4 KB)
I am attaching the requested images
There is indeed something wrong in the way the model constrains where defined for the right hemisphere, but I can't guess what from this limited information.
but of course the lead field vectors are not shown properly
If you want to be able to see something, you should increase the size of the arrows.
But these representations are difficult to understand if you are not very familiar with the question of forward modeling.
I had seen the warnings with the mixed model, but there are two observations based on which I have still preferred using it:
I think this warning is clear eanough: " Mixed head models are mostly indicated for individual subject analysis, and not suitable for group analysis when using individual anatomy for all the subjects".
In the context of group studies: don't use these mixed models.
I strongly recommend you use simpler models: surface or volume.
Thanks a lot
Dear Francois,
Hi again on this topic,
What about exporting the mixed source maps as 4d .nii format (already registered to the subject's own structural) and then mapping them to the MNI template...?
What about exporting the mixed source maps as 4d .nii format (already registered to the subject's own structural) and then mapping them to the MNI template...?
What are you expecting to do with this cumbersome procedure?...