I think I have isolated the issue.
I have started a new protocol.
* yes use protocol's default anatomy
* yes use one channel file per subject
I have kept everything as default (ICBM152)
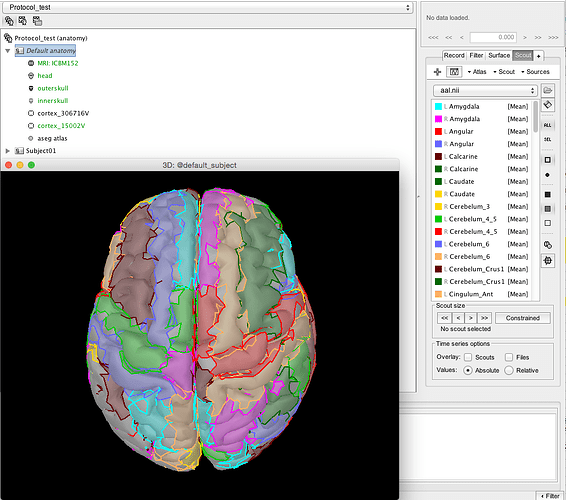
I then load aal.nii.gz (from MRICRON) as a "Volume mask or atlas (MNI space)"
It looks at least like it is in the right orientation to me:
I create a new subject
Import the EEG from an eeglab file
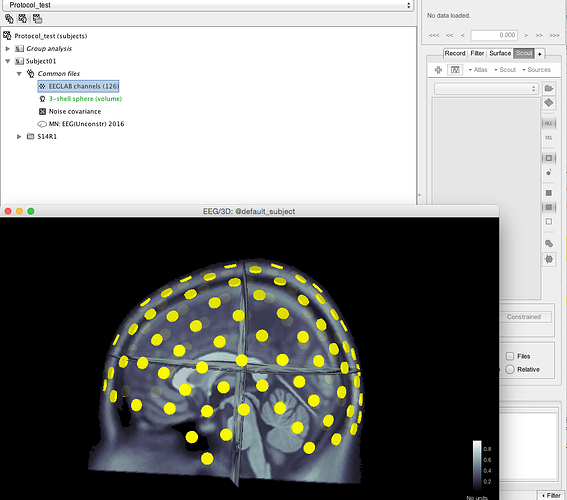
Right click on the EEGlab channels, Add EEG positions, ICBM152, Generic, ASA 10-05 343. It returns that all channels were found
Check the sensors on the MRI:
Create a volume head model (in this case 3-shell sphere, have tried the OpenMEEG)
Create noise covariance, default options
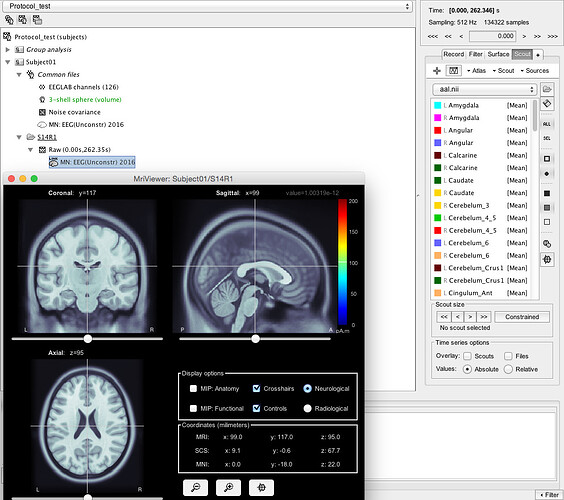
Computed Sources 2016, default options
Double click on the MN: EEG(Unconstr) 2016
Go to Atlas, New Atlas, Volume Scouts
Go to Atas, Add scouts to atlas, "Volume mask or atlas (MNI space)", choose the aal.nii.gz. Scouts are created.
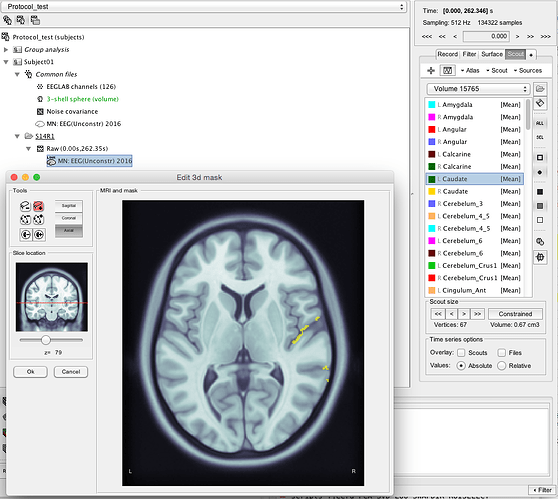
I now have a Volume 15765 atlas with the AAL structures inside.
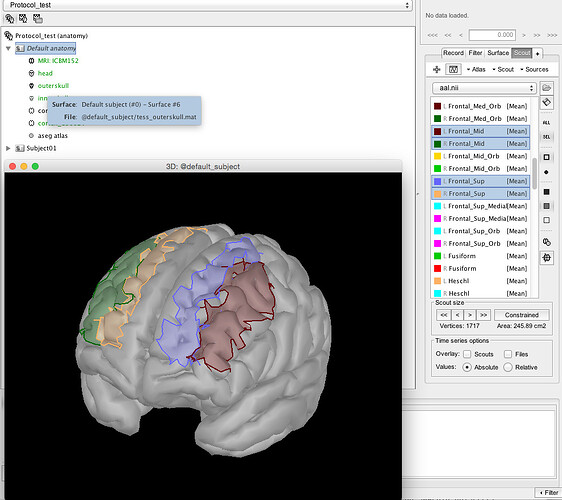
If I click on e.g. the left Caudate and go to Scout, Edit in MRI it looks very wrong: Same with all the other scouts.
I don't understand why the atlas looked aligned when I imported it the first time, but it is wrong when I add them as volume scouts.
Please could you help, I am clearly doing something wrong! If there is any other info or screenshots that I can supply please let me know.
Thank you very much.