Hi Brainstormers,
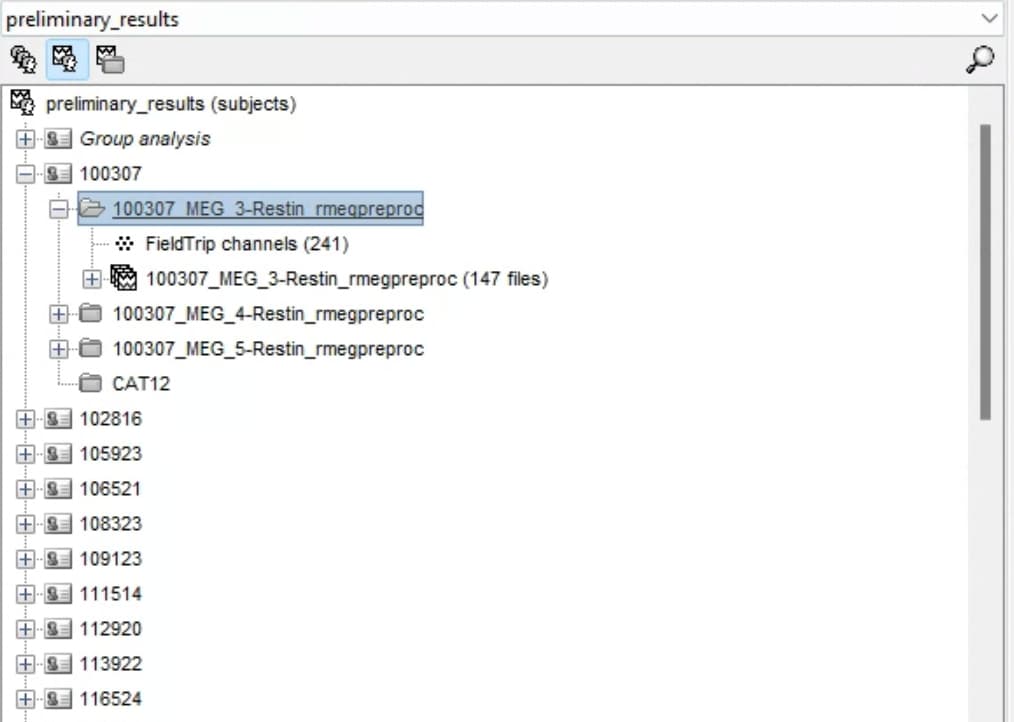
I was trying to use a script to calculate the noise and data covariance for source localization for the HCP preprocessed data. Here is the file structure that I have for my data:
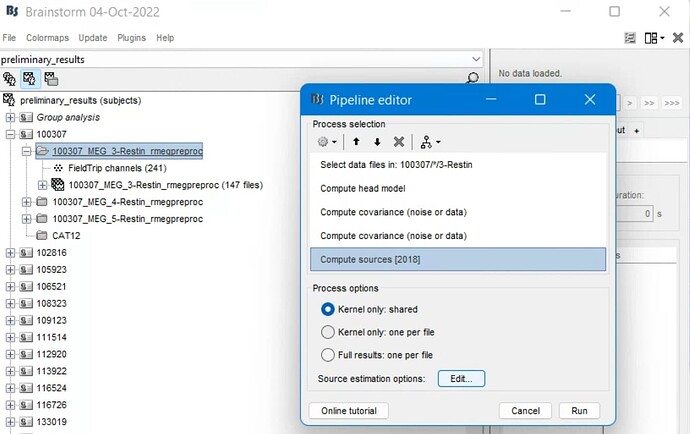
The pipeline that I want to carry out is this:
I wanted to select the files for each session, compute the head model and calculate the data and noise covariance matrix, and then do source localization.
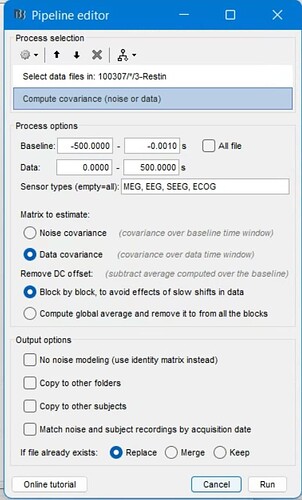
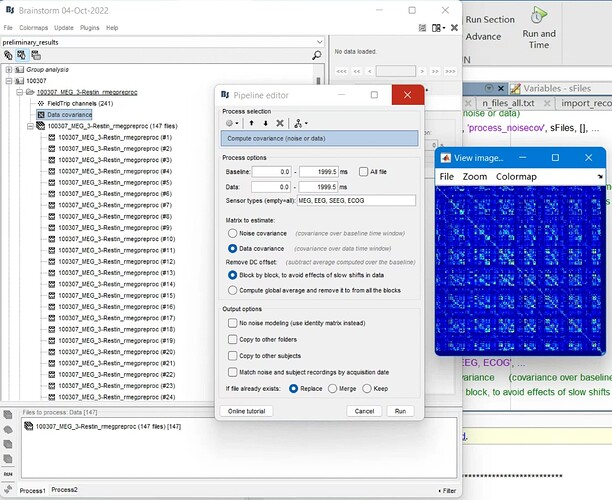
However, the calculation of the covariance matrix seems to behave very strangely when using scripts generated from the pipeline builder. When I select files and then calculate the data covariance matrix:
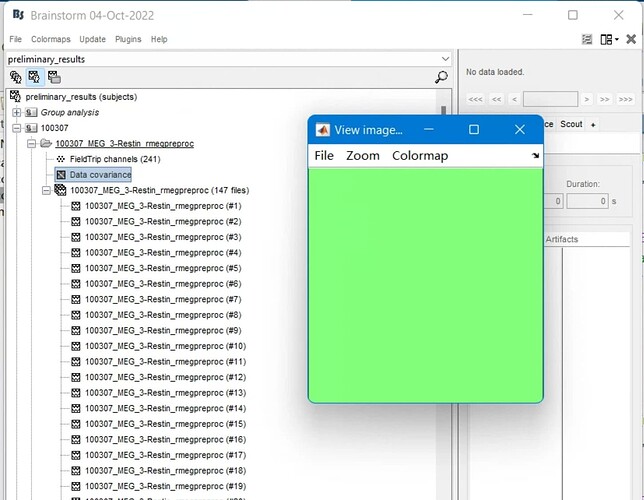
The covariance matrix becomes empty:
But when I drag the files to the process box at the bottom and perform the same process, it gives me the correct matrix:
I also checked the code that I generated from the pipeline and the sFiles output contains all the files I need for the analysis.
% Process: Select data files in: 100307/*/3-Restin
sFiles = bst_process('CallProcess', 'process_select_files_data', sFiles, [], ...
'subjectname', SubjectNames{1}, ...
'condition', '', ...
'tag', tag, ...
'includebad', 0, ...
'includeintra', 0, ...
'includecommon', 0);
% Process: Compute head model
sFiles = bst_process('CallProcess', 'process_headmodel', sFiles, [], ...
'Comment', '', ...
'sourcespace', 1, ... % Cortex surface
'meg', 3, ... % Overlapping spheres
'eeg', 3, ... % OpenMEEG BEM
'ecog', 2, ... % OpenMEEG BEM
'seeg', 2, ... % OpenMEEG BEM
'openmeeg', struct(...
'BemFiles', {{}}, ...
'BemNames', {{'Scalp', 'Skull', 'Brain'}}, ...
'BemCond', [1, 0.0125, 1], ...
'BemSelect', [1, 1, 1], ...
'isAdjoint', 0, ...
'isAdaptative', 1, ...
'isSplit', 0, ...
'SplitLength', 4000), ...
'channelfile', '');
% Process: Compute covariance (noise or data)
sFiles = bst_process('CallProcess', 'process_noisecov', sFiles, [], ...
'baseline', [-500, -0.001], ...
'datatimewindow', [0, 500], ...
'sensortypes', 'MEG, EEG, SEEG, ECOG', ...
'target', 2, ... % Data covariance (covariance over data time window)
'dcoffset', 1, ... % Block by block, to avoid effects of slow shifts in data
'identity', 0, ...
'copycond', 0, ...
'copysubj', 0, ...
'copymatch', 0, ...
'replacefile', 1); % Replace
This problem disrupted my pipeline because the covariance matrix is necessary before source localization. Please let me know if you know what the issue is.
Thank you!