Hi!
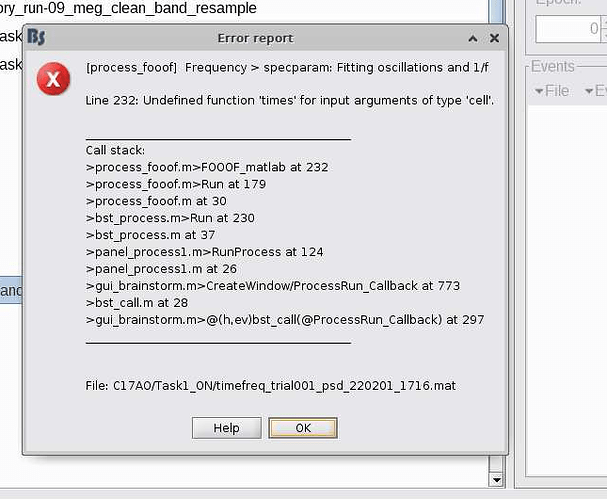
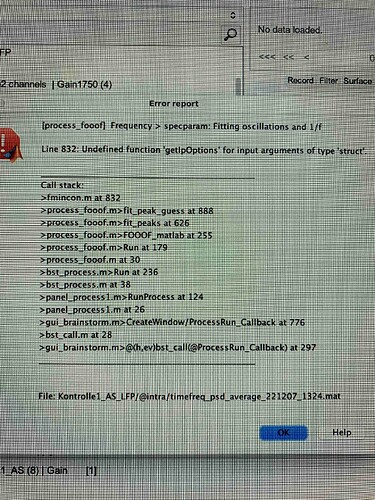
I am trying to run specparam: Fitting oscillations and 1/f on the output from Power spectrum density (Welch), as indicated by the tutorial. I am using the latest version of brainstorm (Jan 31, 2022) and Matlab2021a. I have tried on a few different versions of my data, but consistently get the error shown in the picture. Do you have any suggestions? Thank you!
Keelin
@Luc ?
Hi again. I've tried running PSD followed by specparam: Fitting oscillations and 1/f on data without first applying preprocessing steps to confirm that I haven't created problems through these steps, but I have the same results with PSD working and then getting the following error the specparam function. Do you know what might be causing this error? Thank you!
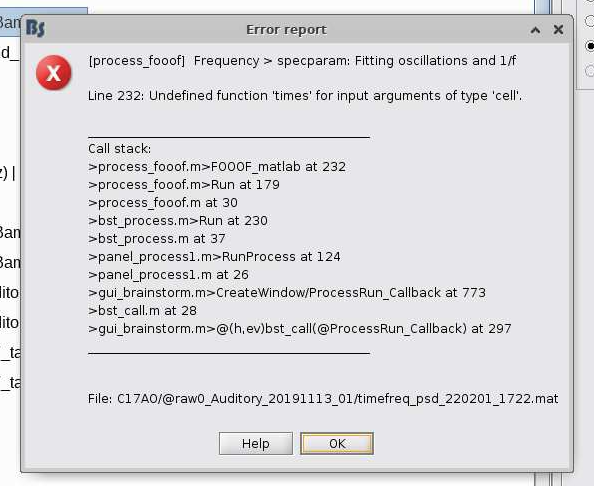
@Luc
Could you please suggest a debugging strategy?
Hi, I am having a similar issue. I used to be able to run specparam just fine before. My initial error is a bit different, but the call stack is very similar:
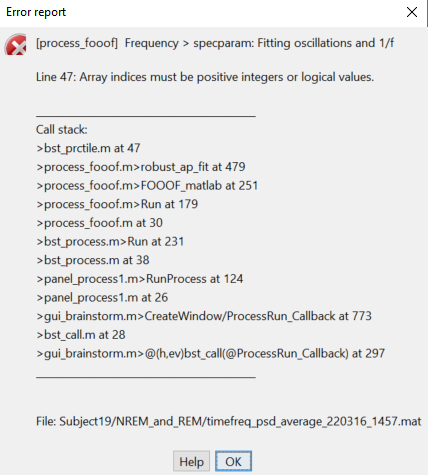
Edit: I've also tried re-running it on a protocol where I had already ran specparam, and I am receiving the same error.
@SoL
Have you executed specparam before on the same file, and it stopped working after a Brainstorm update?
Can you please post screen captures of: 1) the process options, 2) The PSD spectrum plot for your file?
@Luc @Raymundo.Cassani
Any suggestion?
Yes exactly. I was able to run it ~1 month ago, but I have updated Brainstorm since. I initially tried to run it on a different file and saw the error.
To double check, I went to a previous file I've already executed specparam on and received the same error.
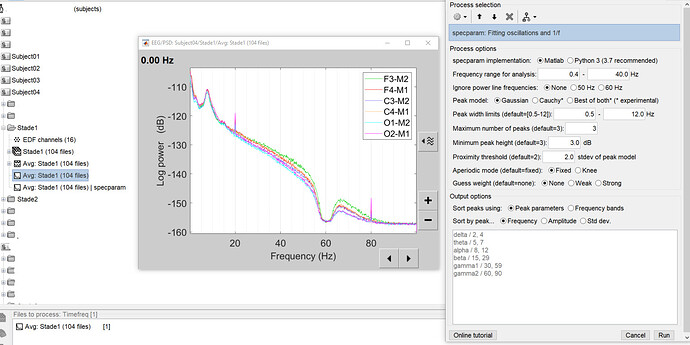
Here is the PSD and the process options:
Thanks.
I will wait for @Luc and @Raymundo.Cassani to reply before trying any clumsy debugging, as they are the experts in this field.
@SoL, could you share the PSD file on which you want to run specparam?
Sure, here:
psd_average_220208_1729-SL.xlsx (98.2 KB)
Hi @SoL, please zip together these 2 files:
- The file PSD to run specparam
(to see where the file is: right-click on it > File > Show in file explorer) - The associated channel.mat file
Upload the zip file somewhere and post the download link here.
Hello,
Thanks !
Soraya
Thank you @SoL
I ran specparam without problems with the shared data. My guess is that your issue is related to the selected parameters for the process.
Please set back the default parameters with the option Reset options
(https://neuroimage.usc.edu/brainstorm/Tutorials/PipelineEditor#Saving_a_pipeline),
and let us know if that solved the issue
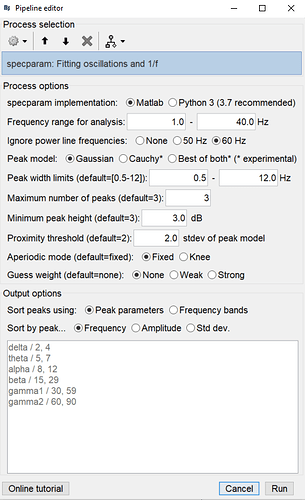
Hi @Raymundo.Cassani , I reset the options however I still get the same error. Here are the parameters it defaulted to
But it's still the same error:

@SoL, thank you for your input
I have found and solved the bug
Just update Brainstorm to get the bug fixed
Git commit: https://github.com/brainstorm-tools/brainstorm3/commit/2141c65f75e7855f9a6506e01b41b72a9afa11fd
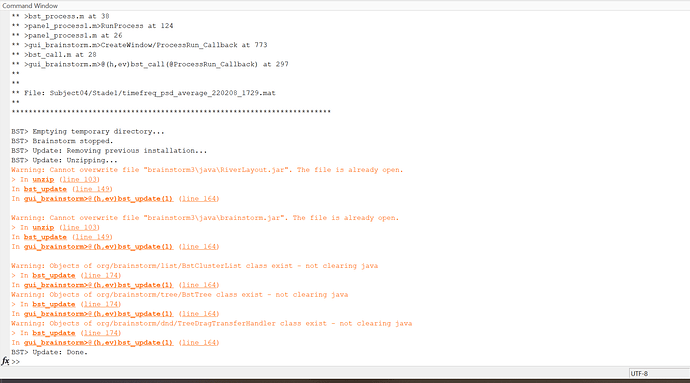
Thank you so much for your help. I have updated Brainstorm-- but I think I am having an error with updating. I'm still getting the same error for specparam, but I think it might just be because my brainstorm is not updating properly. Here is the error I get when updating:
...should I just delete brainstorm and re-install everything?
I assume this shouldn't affect my files or protocol, considering I am not deleting the brainstorm_db folder.
Again thanks for your help
Soraya
It should be ok with those warnings.
Can you try to update again?
Edit: I missed updating the changes to neuroimage.usc.edu server
These warnings should not be a problem, indeed.
But they indicate something that might be a symptom of something else.
I'd recommend you clean up completely your Brainstorm installation if you have more issues:
- Edit your Matlab path, and remove everything that is not related with the Matlab installation itself
- Close Matlab
- Delete ALL the brainstorm3 folders that you have on your computer
- Download a new version of Brainstorm from the Download page of the website: https://neuroimage.usc.edu/bst/download.php
- If you have installed the program correctly (e.g. not setting up your database within the program folder) you wouldn't lose any data or your user preferences (which are in your $HOME/.brainstorm folder)
@Raymundo.Cassani, specparam works now.
@Francois okay I see, thank you for listing out the steps.
Thank you both for your help, have a great day!