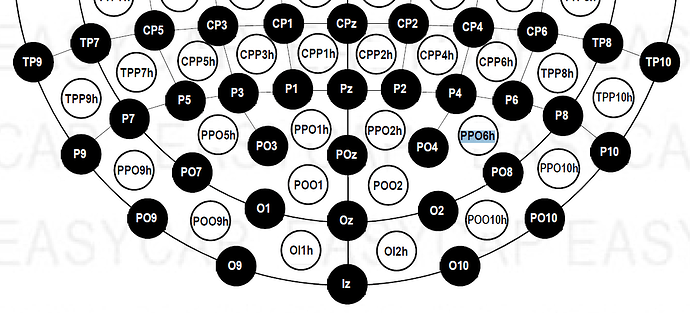
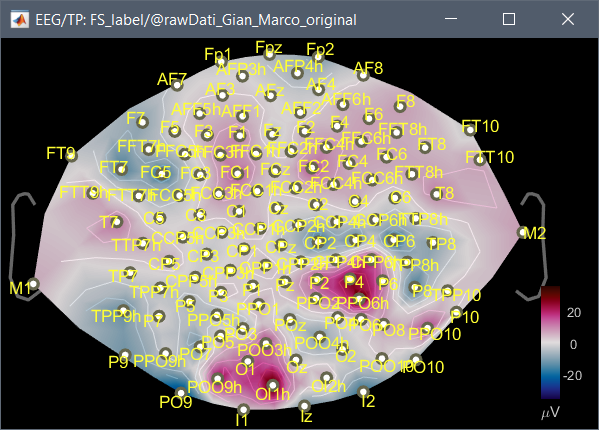
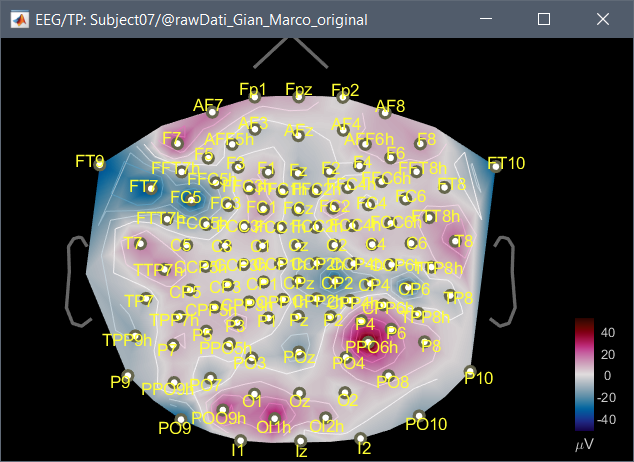
Dear everyone, I have a problem that I'm not solving with a channel location. It is a long story,unfortunately. I recorded EEG signal with micromed system, Easycap 128 channels. Unfortunately, the default channel location in brainstorm cannot be used in my case. I fact when I plot the 2D disc, I have a bad representation on the scalp of the activity. In fact when I imported for the first time the raw signal (.TRC format), I noticed that brainstorm was able to read a chanloc. Even if the values of the channels positions were empty, braistorm was able to read the channel label. I noticed that the channel order in the raw file was different from the one of the default EasyCap 128 chanloc, and moreover some channels were not the same.
I followed two strategies:
- Using the order and the labels of the channels from the raw file, I created a personal chanloc using the xyz position available on internet. Now I have a good 2D Disc representation. Unfortunately some channels in this case are not in the right position, and when I plot the 2D sensor cap the cap has not the right shape, even if the plot on the 2D Disc is correct.
- I tried to modify the default Easycap 128 chanloc of brainstorm, trying to follow the order of the channels in the rawfile. Unfortunately, some channels do not match, therefore I cannot complete the chanloc.
Could you help me to solve this problem?
If you need I could upload 3/4 sec of my signal, so that you can try by yourself to build a chanloc. I can also upload the chanloc I created, so that you can better understand what is my issue.
These data come from an epilepsy population, and I have to reconstruct the source activity of the spikes. Therefore, I cannot approximate the position, as I did, but I need to be as precise as possible, and for this reason I need an additional support from you.
Thanks for your help