Hi everyone,
I'm running into an issue adding eeg channel positions in process.
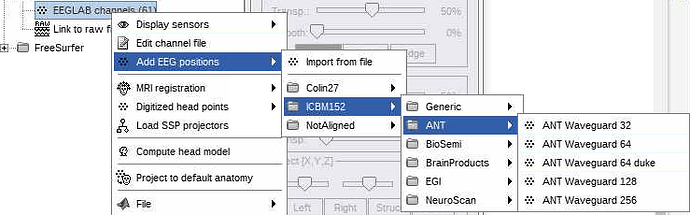
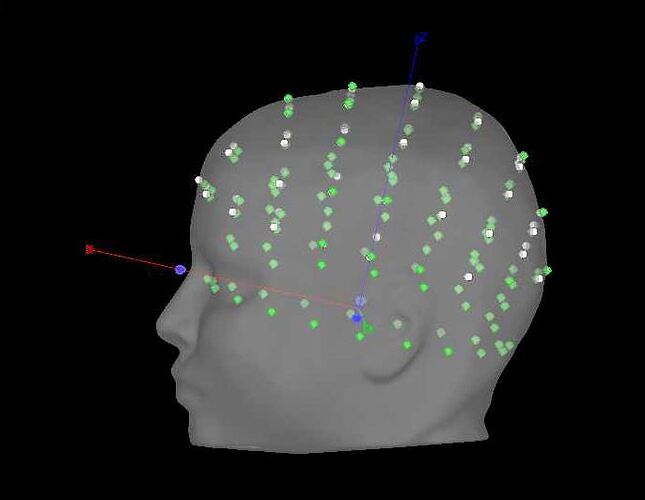
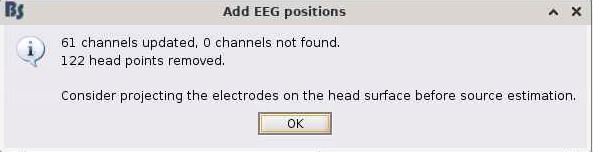
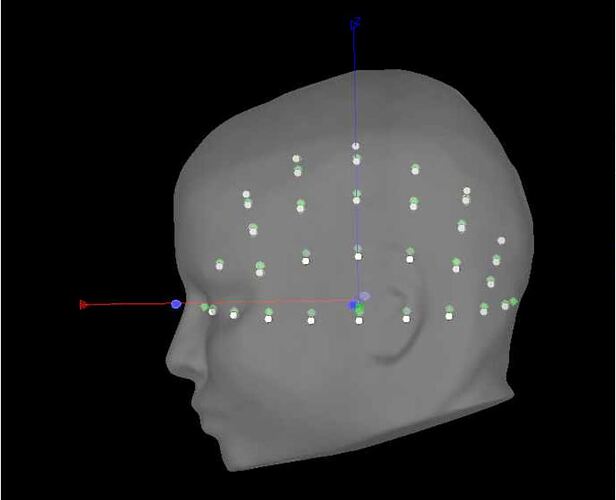
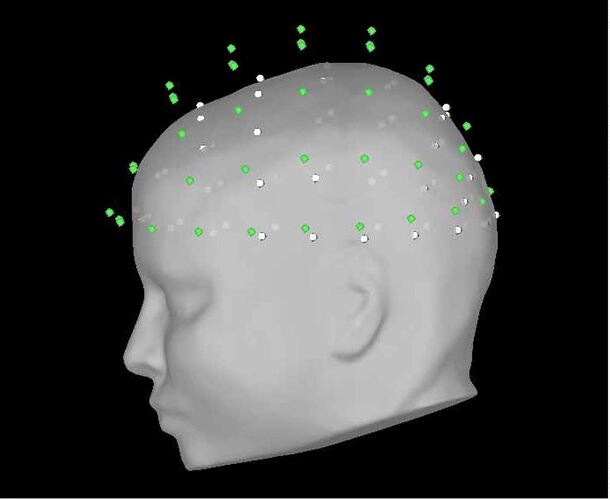
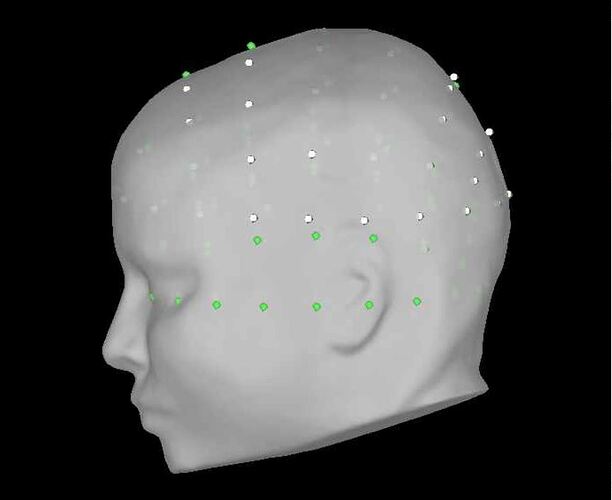
in the GUI, I do the following (image below) and it works great (0% rates between head points and head surface). i select the ant 64 waveguard
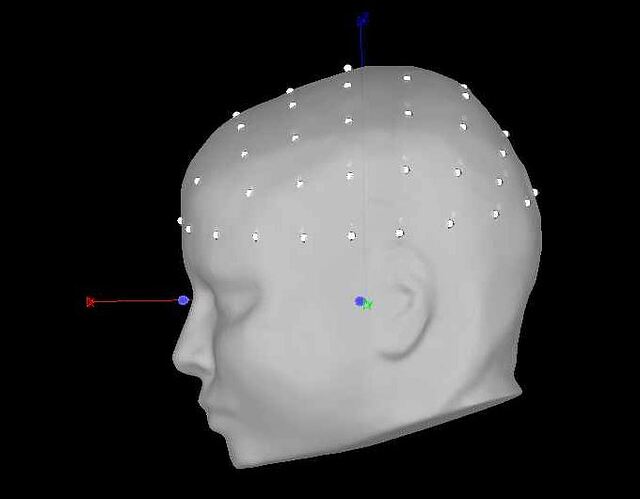
every step before and after positions also works great in 'process'. however, I cannot figure out why when I run add channel positions in process my data goes to 74% rate of >3mm distance between points and surfaces. here is my process script:
% Process: Add EEG positions
sFiles = bst_process('CallProcess', 'process_channel_addloc', sFiles, [], ...
'channelfile', {'', ''}, ...
'usedefault', 'ICBM152: ANT Waveguard 64', ... % ICBM152: ANT Waveguard 64
'fixunits', 0, ...
'vox2ras', 1, ...
'mrifile', {RawFiles{3}, 'BST'}, ...
'fiducials', []);
in the GUI, i am selecting yes to transforming from mni to sensor space (i'm using individual mris). in process, i'm getting the same poor results with and without the vox2ras option.
Curious if anyone has any suggestions. Thank you!!
brian
Brian Kavanaugh, PsyD, ABPP Board Certified Pediatric Neuropsychologist, E. P. Bradley Hospital Assistant Professor, Warren Alpert Medical School of Brown University