Dear all,
I performed a contrast between two conditions at source level using ft_sourcestatistics and to reduce the computational burden, I selected the mean of the Desikan atlas scouts as the entry for the analysis.
The problem is that this pipeline gives as output a file of type Matrix and not a file of type Results (as you get instead from a call to process_test_permutation2p), which I can't reproject to the cortical surface.
Any hints or suggestions to solve this issue?
Many thanks,
Antonio
Hi Antonio
You can probably do something like:
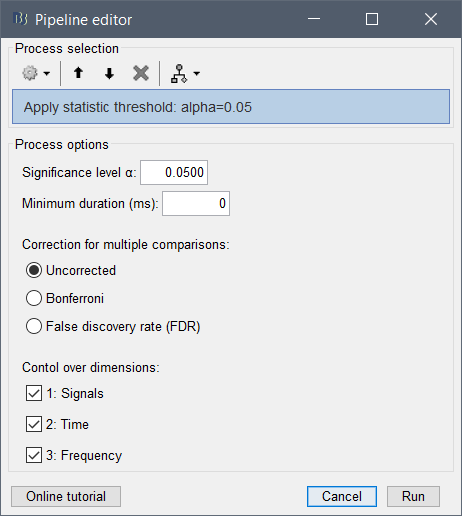
- Call the process "Test > Apply statistics threshold" to create a regular "Matrix" file
- Use this file in input to the process "Simulate > Simulate recordings from scouts" with the option "Save full sources" selected. It would map the scouts onto the cortex (ie. copy each signal value to all the vertices in a scout). The number and order has to be the same in the scouts list (in the options of the process) and in the signals list (in the matrix file), there is no matching done by name.
- It would create also some meaningless "simulated recordings" which you don't care about. Ignore this file and just look at the source file that is linked to this simulated file.
Hi Francois,
thanks for the suggestion. I have just one concern regarding the "Test > Apply statistics threshold". In this process, I don't see the cluster option, so what should I pick to get a cluster-based correction for multiple-comparisons?
In the files produced with the FieldTrip cluster correction, the corrected p-value is already calculated (field pmap in the file). You just need to remove all the values that belong to a cluster that has a p-value that is lower than your desired threshold.
Simply select the option "Uncorrected".
1 Like