Hi Brainstormers,
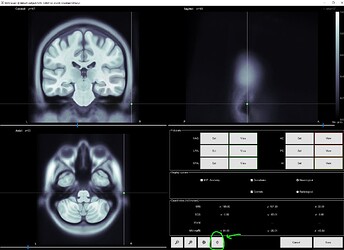
I am wondering if it is possible to project electrodes to the 3D head model generated directly from the standard MNI brain, after clicking this button (green circle):
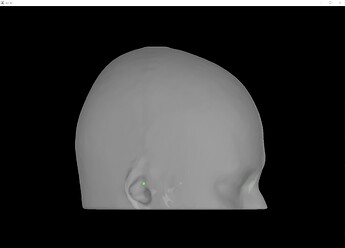
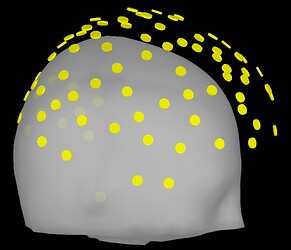
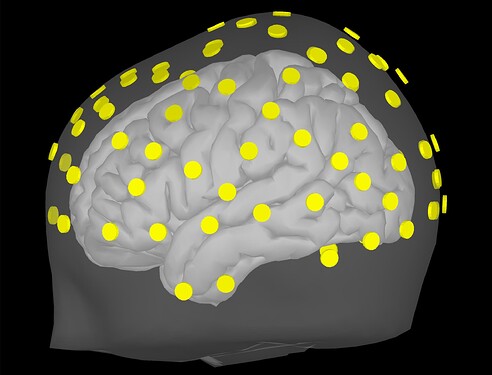
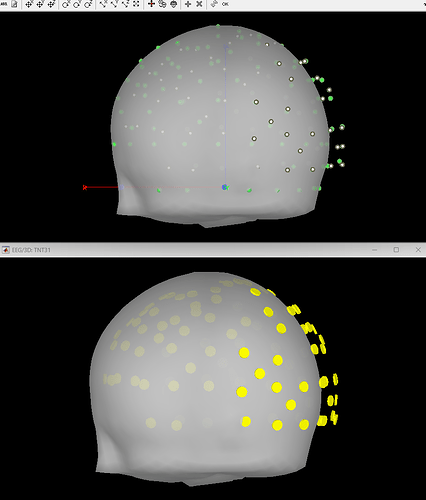
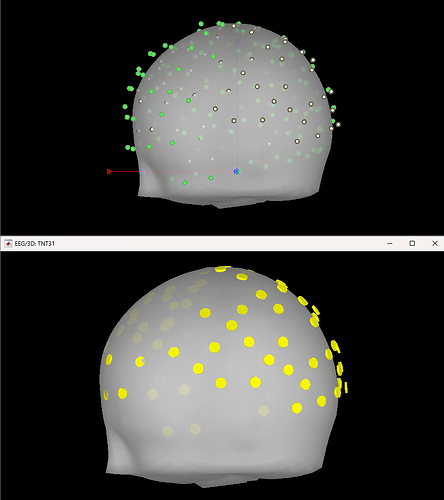
I ask because when projecting to the BEM head model, the fiducials of the MNI brain (which Brainstorm provides) is not very clear to me. I like the visibility of these green dots in this 3D reconstruction:
Maybe there is another way to do this?
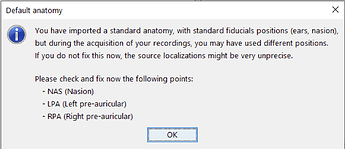
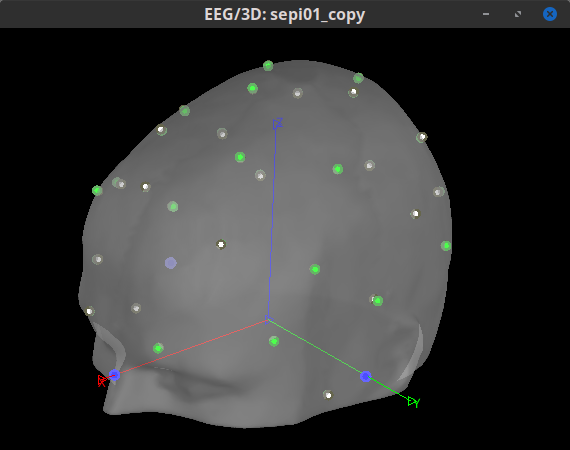
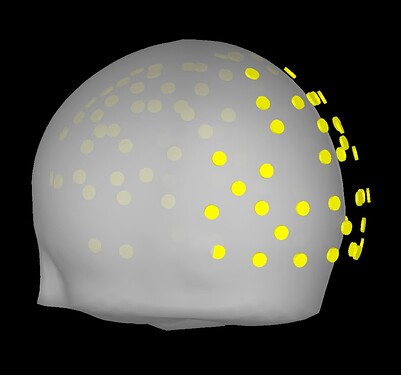
I am inquiring about doing this this because I'd like to ensure that there is an appropriate registration of fiducials between the standard MNI and our CapTrak fiducials, or if I need to edit the standard MNI fiducials. I feel that they are going to be pretty similar, but I want to ensure of this. If there are any differences, I would say that perhaps the RPA position is a little far backwards (caudal) compared to our positions (visible in screenshot 1). I thought we had identical positions until I double checked after seeing these prompts:
How much measurement error (in mm?) between the 2 sets of fiducial points is "acceptable"?
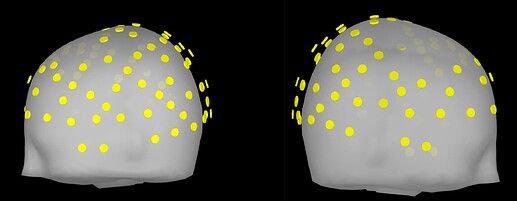
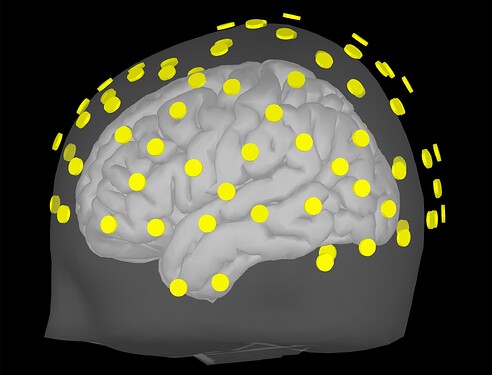
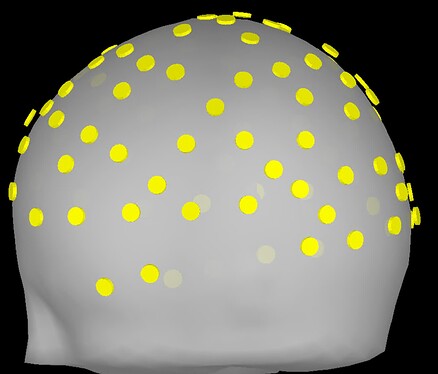
Upon importing CapTrak positions onto the standard MNI brain, I obtained this result (after projecting electrodes to surface), which look completely reasonable to me.
However, I'm wondering if this visual inspection is robust enough to ensure quality EEG-MRI registration with the MNI brain. In looking into File Contents of the CapTrak data, I also notice that the fiducials for CapTrak and MNI brain positions seem slightly off, but this isn't seemingly present in screenshot 5. For example, File Contents of CapTrak give these coordinates (which were in m that i converted to mm):
NAS: [113.2, 0, ~0]
LPA: [13.5, 88.8, 0]
RPA: [13.5, -88.8, ~0]
whereas the MRI viewer of MNI brain told me the following (in mm):
NAS: [101.6, 0, 0]
LPA: [0.66, 83.01, 0]
RPA: [-0.66, -83.01, 0]
While of course there are differences, I am wondering if I am needing to change the positions of the MNI fiducials.
Thank for you for your time. I greatly appreciate your expertise and advice.
Sincerely,
Adam