Hi Kanad,
I have linked the raw data (AUX) first and have performed the necessary pre-processing steps (removal of linear trend, notch filter, baseline correction).
First, the pre-processing of your continuous file should not include any detrending operation or "baseline correction". Those two operations have the same function (the first removes linear trend, the second removes a constant) and are mean to be applied only on imported epochs. It's very difficult to estimate what is the effect of removing the average of the entire recordings....
If you want to remove slow components in your signals (<0.5Hz for instance) at this very early stage of analysis, use a high-pass filter.
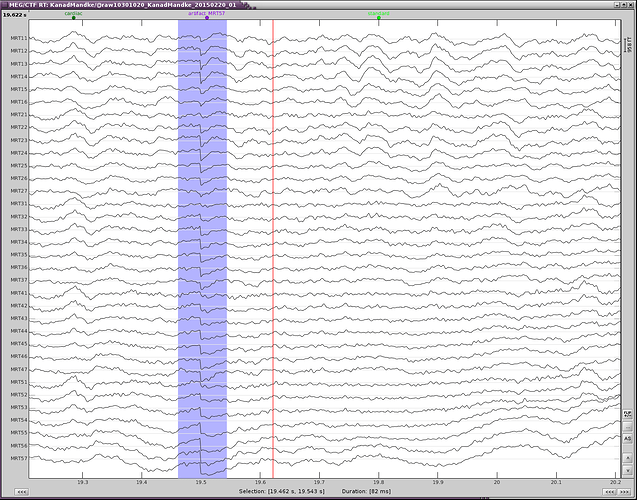
However, whenever I compute SSPs, I don't get a reasonable topography (with PCA) at all. I always have to go through the 'generic' tab and save one averaged component.
Each subject is different and generates different types of artifacts. Brainstorm offers a variety of options, hoping that you will always find one that cleans correctly the artifact.
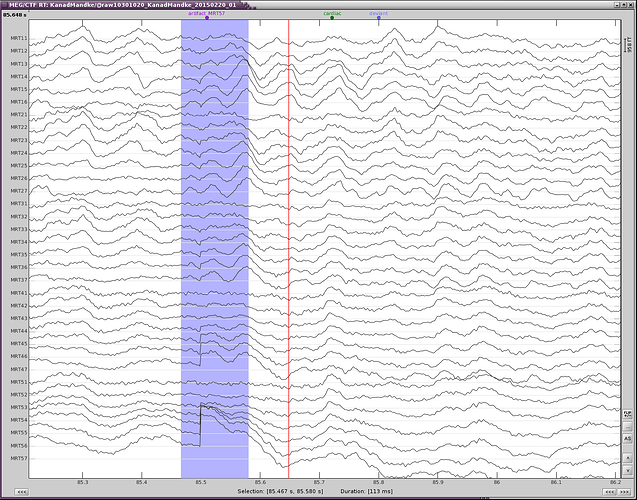
Make sure that you remove the cardiac events occurring at the same time as eye movements, to prevent the first step of cleaning for the heartbeats to parasite the blink correction.
You can also try to uncheck the option "Use existing SSP projectors" or compute them the other way around.
Although that doesn't give me a strong component either.
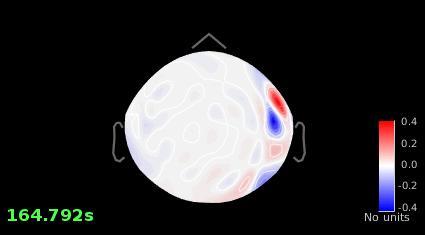
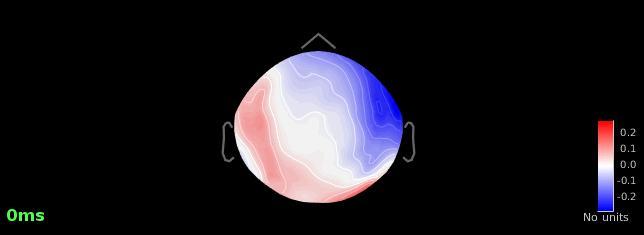
Image #1 looks good, why are you not satisfied with it?
As a consequence, the source maps are still not free of artifacts (activation in the temporal poles, ventral temporal lobes etc).
What kind of experiment is this? Do you have a fixation cross?
If you don't, you would have constant eye movements, almost impossible to remove. The only option would be a high-pass filter high enough to remove them.
If you do have a fixation cross, you may still observe other types of eye movements (saccades or slow movements). You would observe them better on horizontal EOG, and may have to mark them manually and calculate a separate SSP projector.
They are usually easy to see, but might be complicated to correct for...
Can the use of LCMV, instead of dSPM get rid off this problem (is it stable now?)?
A new version of the source estimation framework should be available in the software by the summer, including a LCMV beamformer.
If you have eye movements recorded in your recordings, the inverse solution might help a bit but will not remove them as magically as the SSP.
Secondly, what is the criteria for peak-to-peak artifact rejection that you normally use/recommend in case of evoked responses?
We don't use it. Reviewing manually the recordings is a lot more reliable than any automatic rejection method.
Cheers,
Francois