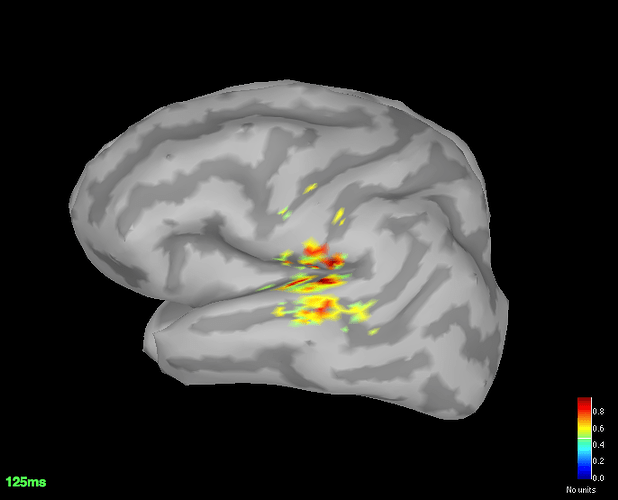
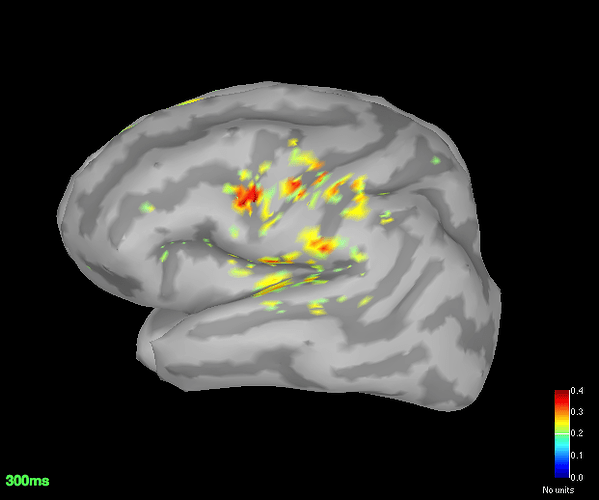
Brainstorm,
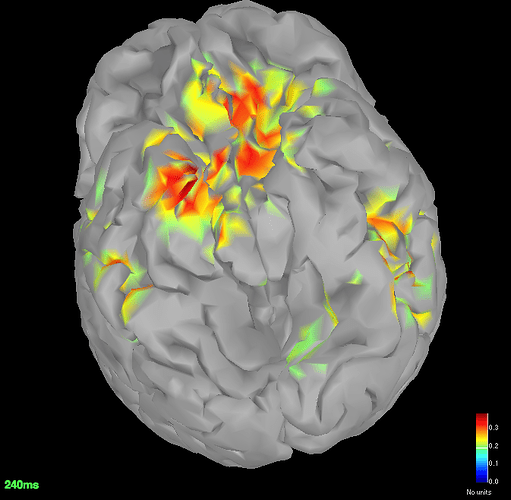
Im projecting the computed sources of one subject onto a Default brain that includes a FreeSurfer Atlas created from all subjects.
My procedure goes like this:
Compute Head Model - overlapping Spheres
Compute Noise Covariance
Compute Sources
Average the sources from all trials
Project Averaged Source file onto the Default anatomy w/ FS atlas.
I have previously done this with the same data set, same participant.
however today im getting this promt
** Error: Line 236: Index exceeds matrix dimensions.
**
** Call stack:
** >bst_project_sources.m at 236
** >bst_call.m at 28
** >tree_callbacks.m>@(h,ev)bst_call(@bst_project_sources,ResultFiles,sDefCortex(iCort).FileName) at 1995***
I run the same procedure on a different participant and works.
Its just one participant who with the same data set, the above procedure worked.
Ive deleted their subject and re-started from scratch (e.g., import FS anatomy folder, review raw MEG file) but i get the same error prompt every time.
I would appreciate your help,
Cheers,
Sean.
Hi Sean,
No I don’t know where this is coming from. In function bst_project_sources, it looks that the function tess_hemisplit is not returning the correct indices (you can explore this problem with the Matlab debugger if you are familiar with it).
Have you made sure that everything is imported properly in the subject anatomy?
For the file cortex_15000V, do you have an atlas “Structures” that contain two entries: “L Cortex” and “R Cortex”?
I can have a look at it if you send me the subject folder:
- remove all the unnecessary files, and keep only the average source file that you are trying to project
- right-click on the subject folder > File > Export subject
- upload the zip file somewhere (dropbox for instance) and send me the link in a separate email
Cheers,
Francois
Hi Francois,
I am getting a similar error message. In my case it is appearing for every subject.
Strange thing is that it worked when I projected the sources on MRI for the first instance then, closed the window and projected the sources on cortex and shifted back to MRI and started getting the error.
Just like Sean, I have computed :
Averaged trials
overlapping spheres
Noise covariance
Computed wMNE
Display on MRI
Error message:
** Error: Line 32: Index exceeds matrix dimensions.
**
** Call stack:
** >tess_interp_mri_data.m at 32
** >panel_surface.m>UpdateOverlayCube at 1815
** >panel_surface.m>UpdateSurfaceColormap at 1522
** >panel_surface.m>UpdateSurfaceData at 1435
** >bst_call.m at 28
** >macro_methodcall.m at 39
** >panel_surface.m at 38
** >bst_figures.m>FireCurrentTimeChanged at 813
** >bst_call.m at 28
** >macro_methodcall.m at 39
** >bst_figures.m at 59
** >panel_time.m>SetCurrentTime at 240
** >bst_call.m at 28
** >macro_methodcall.m at 39
** >panel_time.m at 30
** >figure_timeseries.m>FigureMouseUpCallback at 542
**
Secondly, (when it worked the first instance) even though the head model seems appropriate the activation is seen (concentrated) at the base of the brain when the experimental task is auditory oddball. With wMNE, dSPM & LCMV.
If this error is solved I might be able to post pictures so to give you a better idea.
Best,
Kanad
Hi Kanad,
This is not the same type of error as the one you are referring to.
In Sean’s case, it was due to the projection to the default anatomy (Colin27).
In your case it’s just the re-interpolation of the surface into the volume, it is probably a simpler problem.
The first thing you could try is to remove all the existing interpolations from the surface files.
In the anatomy view, right-click on the cortex file > Remove interpolations.
Let me know if you the error stays.
Have you tried just re-importing everything for this subject? Sometimes a manipulation error at some point can lead to strange errors.
Have you calculated the sources on the surface (like in the introduction tutorials) or in the volume (like in this tutorial: http://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource)?
If you are interested in unconstrained positions in the entire volume, you might be interested in the second option.
If you get errors or weird results, try first on a surface-based approach: because there are a lot more users for those methods, the code has been debugged a lot more over the years.
If you don’t manage to solve your problems, please post a few screen captures (showing the structure of your database, the MRI/sensor registration, the ERP…)
Alternatively, if you have subject that is not too big, you can send it to me: right-click on the subject > File > Export subject. Then upload the .zip file somewhere and send me the link by email (click on my username on this forum).
Cheers,
Francois
Hi Francois,
Thanks for clarifying.
I had already tried the remove interpolations options but the error was persistent. I was trying to avoid re-importing everything but I’ll just give that a go. Hope that sorts it out.
I am interested in the unconstrained positions based on the entire MR volume (second tutorial) but as you have suggested I’ll just try a surfaced based option, if that doesnt work i’ll send you an email.
Thanks again.
Kanad
Hi Kanad,
The MEG-MRI registration and the evoked responses look really good, you should be able to get correct source maps.
- How do you use the .pos file? If it has the correct format (same as in the dataset of introduction tutorial), you just have to copy it inside the .ds folder and it should be used automatically. Don’t try to add it manually after linking the .ds folder to your database.
- Can you post a screen capture that shows the head shape (green points) loaded from this .pos file
- Where is this head surface coming from? The holes in it are probably preventing the automatic registration to work. Fix this before you try to re-load your data with the .pos file. Any particular reason for which you don’t use the head surface generated by Brainstorm?
- Is your version of Brainstorm up-to-date (less than a month old)?
- At the MNI, we don’t really use the normalized sLORETA solution. We prefer to use either Z-scored wMNE maps (or dSPM). We cannot provide much support regarding those results. Please post screen captures of your MNE and Z-scored MNE maps instead.
- Are you sure your noise covariance matrix has been correctly estimated. A badly defined baseline / noise covariance is one common source of non-sense source maps. If the segments of recordings you used for estimating the noise covariance contain some of the brain activity of interest (in your case some auditory activity), the auditory cortex may tend to be excluded from your source reconstruction. This is why we always recommend to use a noise covariance calculated from empty room recordings, or resting state recordings.
- I am currently working on an auditory tutorial, this may help you as well (the source part is not finished yet, but will be in the next few days):
http://neuroimage.usc.edu/brainstorm/Tutorials/Auditory
Cheers,
Francois
- I initially had copied the .pos in the .ds folder before importing the data and got these results. After which I removed the file and integrated that through digitised head points --> add points (though I did notice, .pos is under EEG) which still gave the same results. For the previous post I completely removed the .pos file and computed the headmodels/mri alignment only based on fiducial, which still has given quite a good fit.
You can either leave the .pos file in the .ds folder (and say YES to the automatic registration), or just completely ignore it.
I would not add it after manually, as it doesn't do exactly the same thing.
- Unfortunately, the stylus that we have is rather sluggish and hence it sometimes records unintended data points, 2 outliers. But Im sure those will be removed when refining the headshape. However, I have not used the digitised headshape (for the previous set of screenshots I did not use the .pos file). How do you suggest I check if the holes are really interfering? If then, how do I fix those?
This fitting process could be a bit biased by the outliers, you should remove them from the pos file manually.
They are easy to find, they are the two ones with the extreme values in X coordinates (press CTRL+A to see the axes)
- We usually generate surfaces using some in house packages but on this occasion I have used BrainVisa (I dont have separated MR scans of the brain for all participants). I have imported those surfaces into BS and down sampled them before merging the surfaces (as mentioned in the tutorial).
I recommend you import the BrainVISA / FreeSurfer segmentation results with the menu "Import anatomy folder", as indicated in those tutorials:
http://neuroimage.usc.edu/brainstorm/Tutorials/SegBrainVisa
http://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer
- Have you correctly removed the blinks from your recordings?
- I was using the pre-stimulus interval to calculate the noise covariance. As per your recommendation, I have used an empty room recording to compute the covariance matrix and used that for further processing. This has improved the results slightly but the basal activity is still there. Although misplaced, the activation has now moved laterally.
Note that the noise recordings have to come from the same day.
- I still think the problem is somewhere in the headshape/co-registeration.
The registration looks good, the helmet is correctly placed on the subject's head. If you have a registration problem here, it would not influence the results more than for a few millimeters in spatial accuracy.
I would tend to think it is more due to the modeling itself (noise covariance and/or inverse model).
Hi Kanad,
It is difficult to evaluate if the data was properly imported just by looking at your Brainstorm database.
Could you send me your original recordings as well?
(Empty room recordings + subject recordings + anatomy)
Thanks,
Francois
Hi Francois,
Thank you so much for your detailed reply.
This will definitely help me.
This specific recording is from a MEG naive subject and its one of the first measurements that I did. After I realised the shortcomings from this recording I have optimised the protocol even further. fixation cross, additional electrodes etc.,
-
Yes I am re-calculating all the averages in Brainstorm from the .ds (epoched) file.
-
The EEG channels in this case do not have anything. I’ll be adding Cz & Pz later.
-
should I use *.ds, convert it to continuous in Brainstorm and then import it like in the tutorial or use _AUX.ds instead?
-
yes, MLT23 has been marked out.
-
I had removed some eye blinks but maybe not all. I’ll try again with a stricter criterion.
-
Yes, bipolar electrodes will be added in the future recordings. Thanks for recommending that.
Just to clarify what I understand, so once I link the raw file, i’ll go through the trials and mark the eye blinks and group them together. From there on will i be able to link the custom group in the SSP pipeline without the EOG channel? or should I identify the blinks on one particular MEG channel and use that in the SSP pipeline?
Beyond this point, if needed I’ll add a high-pass filter.
Sorry for bothering you again, Im quite new to this area and still exploring everything.
Thanks,
Kanad
Hello
- should I use *.ds, convert it to continuous in Brainstorm and then import it like in the tutorial or use _AUX.ds instead?
We use preferentially the AUX files, but if you set correctly the acquisition to have a continuous file in the regular .ds, it doesn't matter.
For example, the auditory tutorial uses "epoched" datasets.
Just to clarify what I understand, so once I link the raw file, i'll go through the trials and mark the eye blinks and group them together. From there on will i be able to link the custom group in the SSP pipeline without the EOG channel? or should I identify the blinks on one particular MEG channel and use that in the SSP pipeline?
You could do both: mark the events manually, or try to detect them on one frontal MEG channel where the blinks are very strong.
For the SSP calculation, all you need is a category of events that points at one precise artifact, it doesn't matter how this events have been marked or detected.
One the subject I looked at, given the amount of eye movements you have, you probably didn't have a fixation cross.
You can try with SSPs, but you may need a high-pass filter if it doesn't remove the artifacts correctly.
Cheers,
Francois
Hi Francois,
I have followed every step according to what you have suggested so far and what is mentioned in the tutorials but I am still getting predominant basal activation.
-
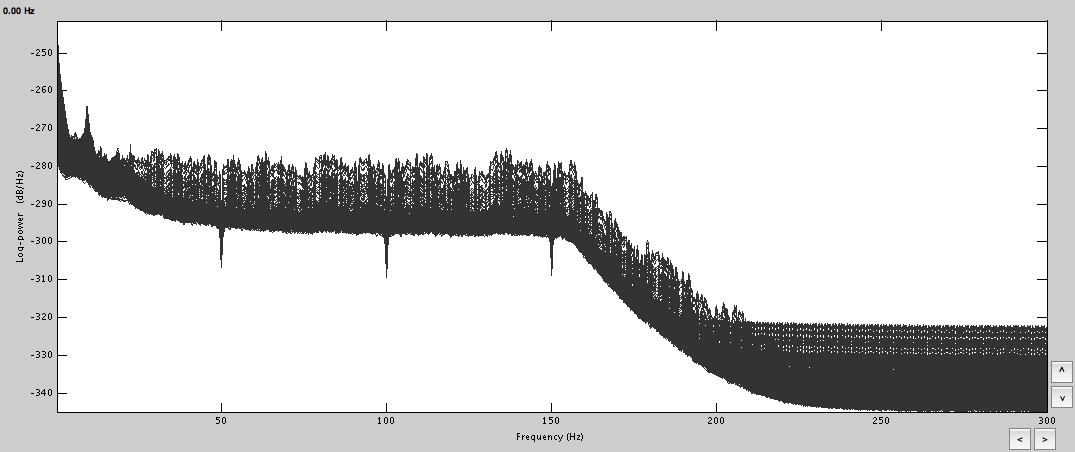
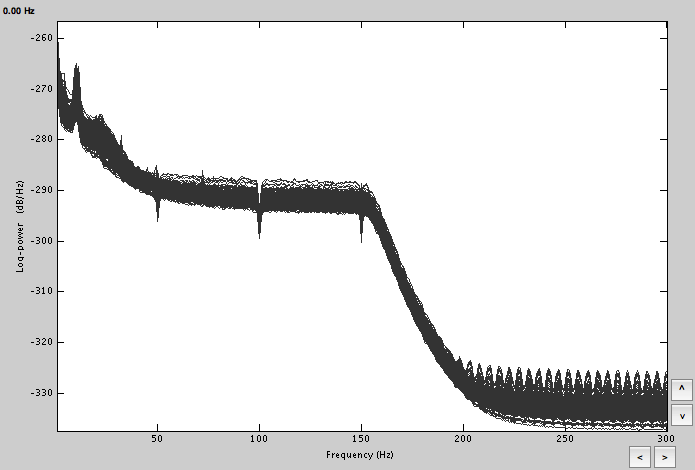
To start with, I linked the AUX file and did the spectral evaluation to remove 50Hz harmonics, mark some bad channels and segments.
-
Manually marked eye blinks, saccades and heart beats across all trials.
Computed the SSPs according to the SSP tutorial. Cardiac first, blinks later and then a generic SSP for saccades.
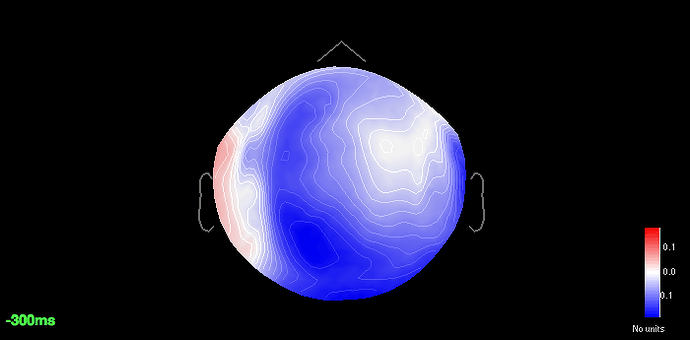
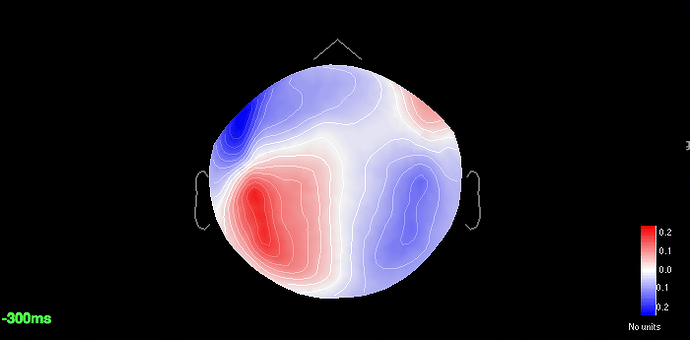
All the major components are 12%, 19% and 18% respectively. Specifically for saccades if I add a filter of 1Hz-2.5Hz the component weight increases upto 31% but that affects the p3 amplitude greatly. So I resorted to the 18% component. For blinks and saccades the sensor cap shows a strong dipolar pattern for frontal regions.
-
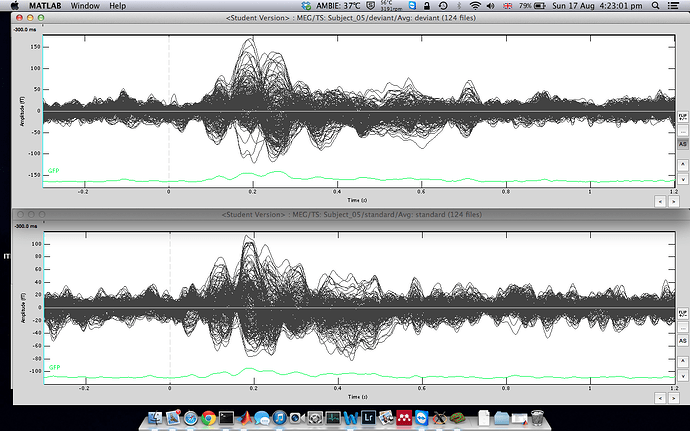
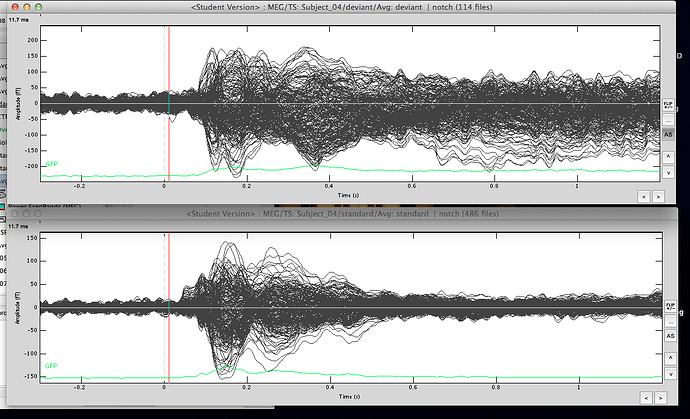
The averaged waveforms for deviant & standard
-
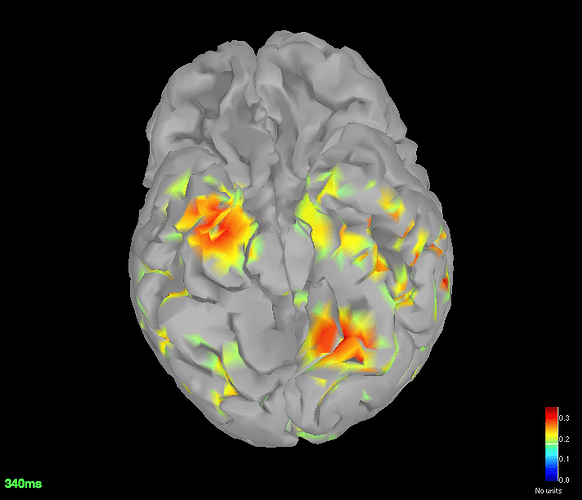
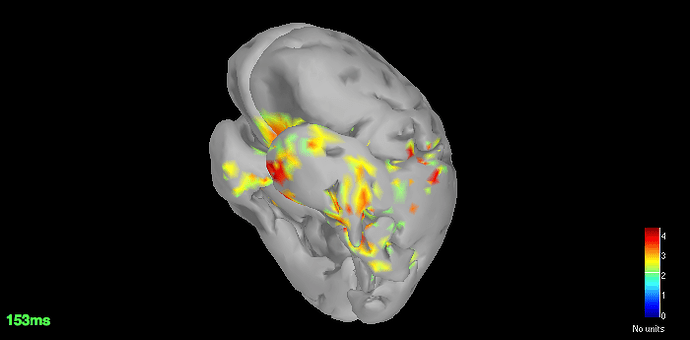
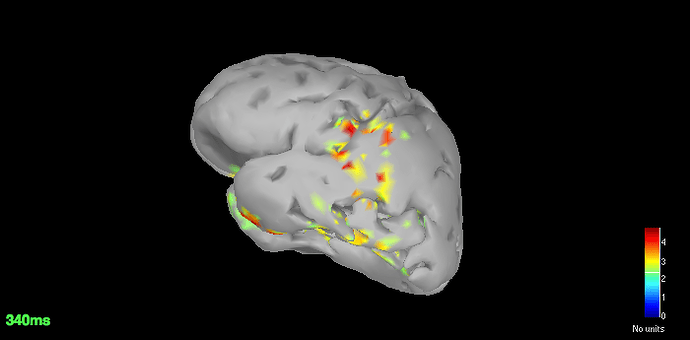
Source space (dSPM) averages for deviant and standard
-
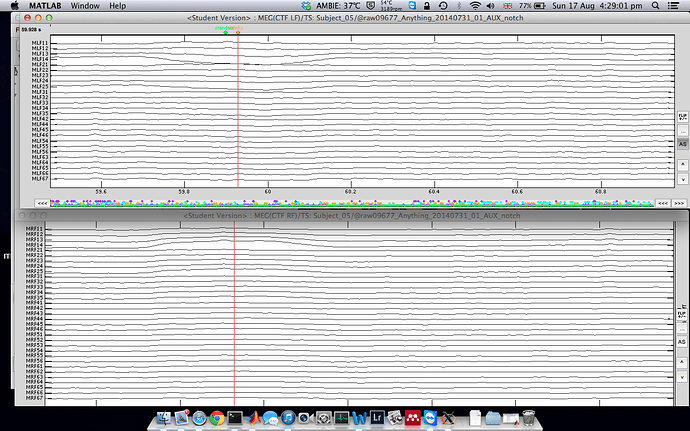
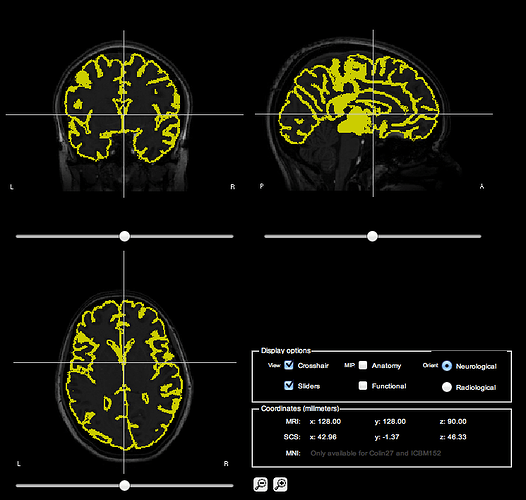
MRI Co-reg for your reference
Since you got good localisation for N100, I am quite sure I have made a mistake somewhere but can't seem to isolate that.
Please let me know.
Best,
Kanad
Hi Francois,
Thanks for your reply.
The SSP that I posted earlier, blinks & saccades though had same pattern had different component weights. I have always used ''use existing SSP projectors''
Here are the SSPs remarked:
Blinks 11%
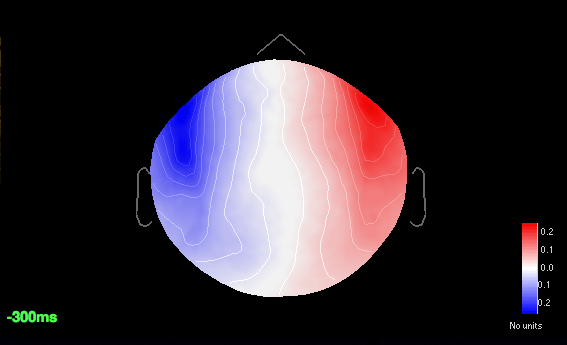
Cardiac 9%
Saccadic 8%
-
Sorry if I didn't make it clear at the start, but it is an eye open experiment.
Since it was the first run testing the entire setup, I didn't have a fixation cross. But yes, I have added that now with additional electrodes for future measurements.
-
I have processed the dataset in both continuous and epoched files.
With _AUX.ds (previous screenshots) and *.ds today.
Here is what I have done so far:
Linked the epoched (*.ds) file, marked the events.
Computed the SSPs, blinks first, saccades later. Imported the recording as standard & deviant.
Computed FFT for both and marked the bad channels. Removed the power line contamination. Based on those, got the averages, which do look a lot cleaner.
FFT:
Averages: deviant & standard
Got the dSPM for both the conditions.
The localisation has slightly improved, in terms that it at least is now showing us activation in some what reasonable location. However, the eye related artefacts are still present. Interestingly, the activation seems to have 'lateralised' to the left side hemisphere.
Standard average
Deviant average
Lastly, the way I understand the procedure of generating dSPM is that we compute it for every trial and average it in the source space before displaying (induced responses?).
So is there a way to construct sources directly for the cleaner averaged waveform? This wont be as accurate as the method described in the tutorial but wont it get rid off the eye artefacts as those wont be time locked?
Thanks,
Kanad
Hi Kanad,
-
Your averages look a lot cleaner. But all this mess at the end of the deviant average may indicate that you have a lot of eye movements left. Check your trials one by one, at this point you have no other option. The ocular artifacts (blinks or movements) are not time locked but they are huge, the amplitudes are so high that you need many hundreds of trials to have them disappear in the average.
-
Your blink component looks ok, but I’m not really sure about the two others. The cardiac component may have some auditory or ocular activity in it. The saccade one contains some auditory activity. Try to refine your computation.
-
After you calculate all your SSP projectors, review all the recordings manually again and mark as bad all the segments that still look noisy (select time segment and CTRL+B or right-click > Mark segment as bad). You still have some strong artifacts to remove.
-
I see your averages are called “deviant | notch” and “average | notch”. This means that you applied a notch filter on the imported epochs. This is not a recommended operation. The edge effects of any frequency filter can be important, we only apply them on long continuous signals. If you have the AUX files, you should work only with them and do all the pre-processing and artifact cleaning on the entire sessions.
-
Your cortex surface looks bad. What did you use to generate this? I would recommend you try with FreeSurfer or BrainSuite instead (check the tutorials online).
-
About the averages: within one run (one channel file), you can compute a shared inversion kernel, calculate the average of the recordings per condition and use the source link of this averages. If the position of the head does not change: sources(average)=average(sources). Then if you want to average across runs or subjects, you need to average in source space for accurate results.
Good luck, you’re almost there =)
Francois
Hi Francois,
The SSPs and high pass filter worked like a charm. The filter has of course reduced the p3 response significantly but atleast Im getting fairly good localisation.
The last surface was generated using BrainVisa, even I noticed how bad it was, i'll switch to FreeSurfer. Until then, this is on Colin27.
Avg standard
Avg deviant
Thanks a lot for your quick replies. If you're ever in England, your drink is on me. =)
Cheers,
Kanad
Hi Kanad,
Glad we could help. You can also thank Beth Bock, the MEG engineer at McGill, she’s the local expert in artifact cleaning and she guided me for all those advices.
I take the drink offer seriously: we might be coming to the UK for a Brainstorm training some time next year =)
Francois
Brilliant. Looking forward to some Brainstorming in UK. =)
Thanks both of you! I’ll keep an eye for your updates.
Just a quick question, is there a way to convert the scout time series to a PSD plot?
Best,
Kanad
Do you mean: calculate the PSD for a signal you extract from a scout?
Yes, save the scout time series (process Extract > Scouts time series), then drag and drop the scout file to Process1 and run process Frequency > PSD.
To do it properly for one scout, you may want to calculate the PSD on all the source signals (set the scout function to ALL), then calculate the PSD, and finally average the power for each scout (process Average > Average rows).
Francois