Hello Brainstorm community,
I am trying to export the following preprocessed-raw-continuous recording consisting of 15 EEG channels, 2 EOG channels (EXG1 and EXG2) and 3 EMG channels (EXG3-5) into pdf pages of 30-sec epochs for sleep stage scoring and list some related questions. At the moment the EXG1-5 channels are classified within Brainstorm channel editing as EEG but they are actually EOG and EMG channels. This is so that EEG-EOG-EMG channels can be viewed on one page without opening a new window for each channel type.
-
I know that there is a way to specify channel type (as in EEG, EOG and EMG) using the following process "Import > Channel file > Set channels type".
However is there a Brainstorm process to change the NAME of the channel? I need to change the name of the EOG and EMG channels and was wondering if there is a fast way to do this (the EXG1-5 channels into something like left and right eye and chin muscle). -
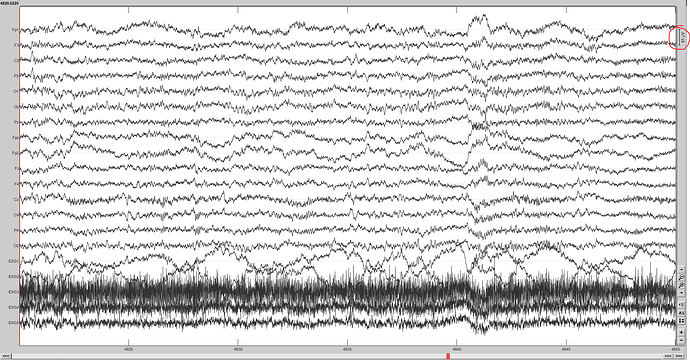
At the moment you will see at the top right-hand corner (circled in red) that the channels are being viewed at 51uV. Is there a way to specify this differently for each channel?
As in EEG channels get 50uV, EOG and EMG at 25 uV. -
What would be the best way to export this continuous recording into 30-sec epochs?
I've tried "Import in database" specifying that the recording be split into 30-sec epochs.
This does not work fully as the epochs do not contain EOG and EMG (which I already specified to be EEG in the continuous recording). They do not appear because through the "Import in database" process, Brainstorm automatically reclassified them as EOG-NO-LOC.
Therefore how would you overcome this problem?
And subsequently, what would be the best way to export all 30-sec epochs into one file so that the end result is one file which I can flick through like a book so that each page equals a 30-sec epoch.
Many thanks in advance.