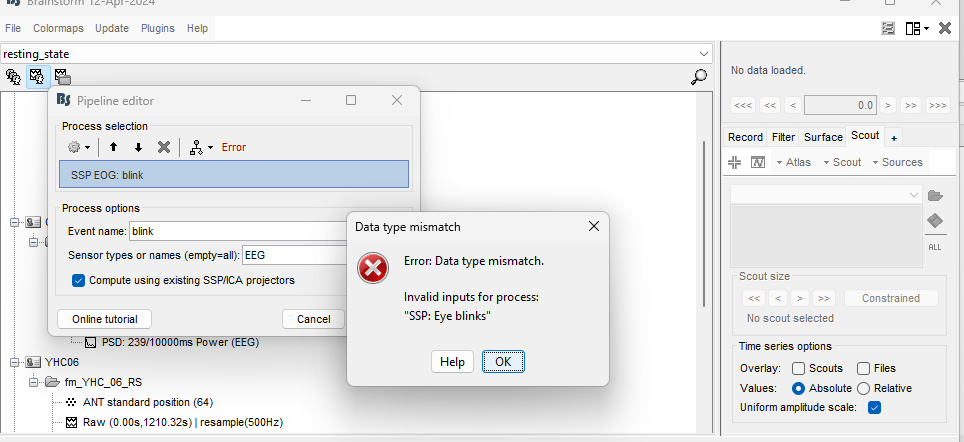
hello i'm working on removing artifacts ( eye blinks, musclecontraction), but i keep getting error on my screen. does anyone know what i might be doing wrong? i've already looked at brainstrom's tutorials but i keep coming up with error. can anyone help me with this?
The process SSP: Eye blinks requires the input to be a raw (not-imported-yet) data (![]() )
)
Could it be that you are trying to run this process with imported data (icon without the RAW indicator)?
Hello I have now created a link to the raw file. So now no more error appears, but it takes very long before I get result. Is this normal? It says all the time on my screen. " calling external function: picard ()..."
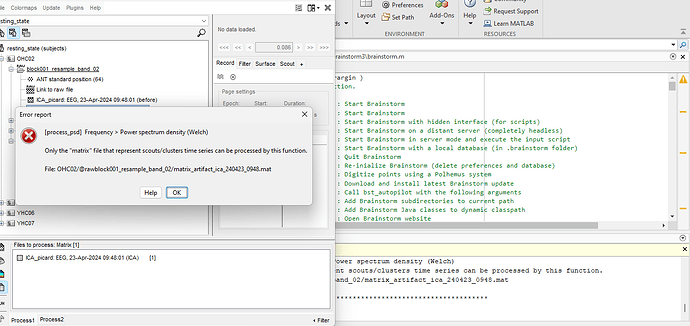
update: After waiting a long time, I finally managed to get an image. i now want to render this as a function of frequency. so as a PSD. but i keep getting an error. how can i solve it? I also have a file called " ica picard : eeg ... ( before)" that was created when running the artifact pre-processes. but I assume this is my old image? the PSD does work on that.
again an update ![]() I have converted the data to raw data and did manage to get a PSD. my question now is, how can I know that my artifacts are really gone? i had also before i wanted to remove the artifacts also applied and resampled a bandpass filter. does it remain present if i worked with the raw data on which i resampled and applied a band pass filter? or do you need to remove the artifacts first and only then apply the bandpass filter.
I have converted the data to raw data and did manage to get a PSD. my question now is, how can I know that my artifacts are really gone? i had also before i wanted to remove the artifacts also applied and resampled a bandpass filter. does it remain present if i worked with the raw data on which i resampled and applied a band pass filter? or do you need to remove the artifacts first and only then apply the bandpass filter.
Hi @Bachelorproef you should read the below two tutorials for topics regarding artifacts.
https://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsDetect
https://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsSsp
If you did artifact rejection > bandpass filter and/or resample, the artifact rejections should be there still. You may need to select process entire fill at once for files with SSP (if you did artifact rejection with SSP).
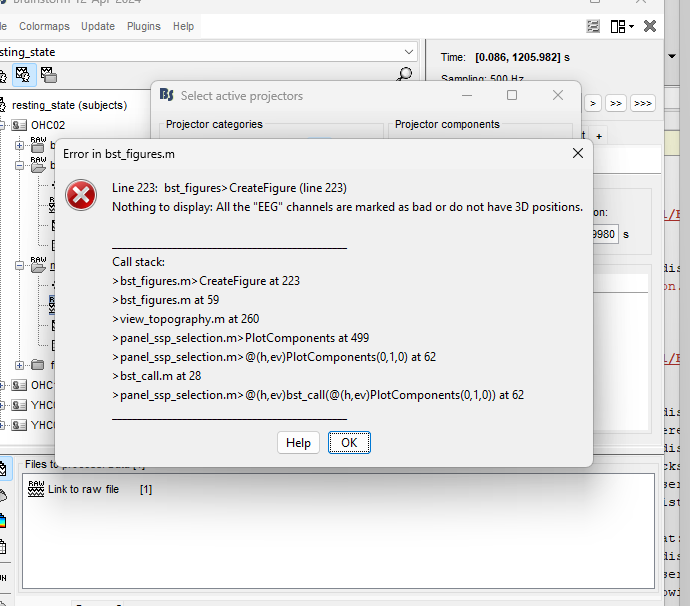
i ran an ICA and now i want to view the different components but i keep getting an error. how can i solve this?
It seems your EEG channels do not have locations. Thus, it is not possible to plot the IC weights in a topoplot.
If you are new to Brainstorm, we recommend you start by following the introduction tutorials first (section "Get started" on the tutorial page), using the example dataset that is provided.
https://neuroimage.usc.edu/brainstorm/Tutorials