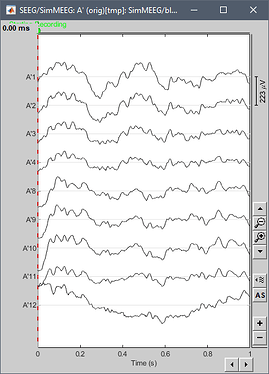
Hello,
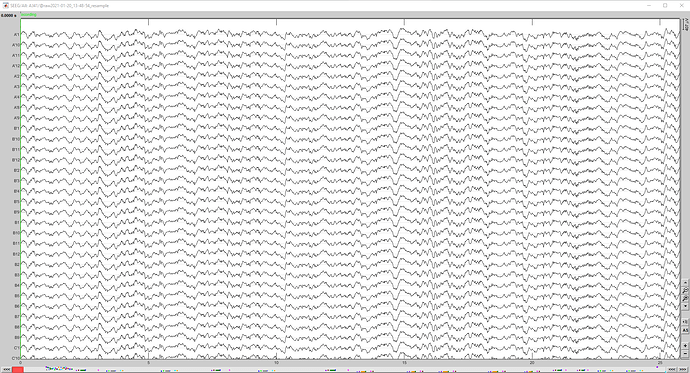
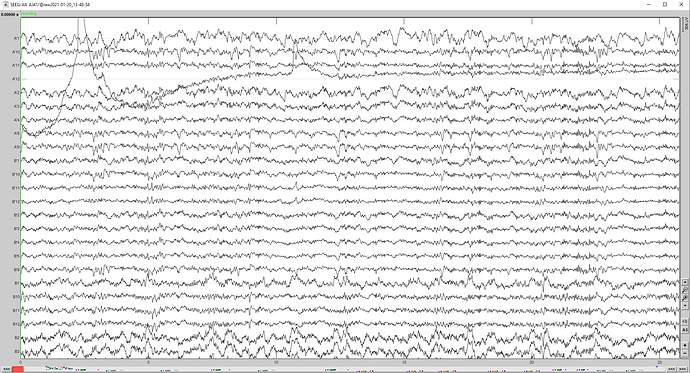
I have a problem when I try to resample my sEEG data. When I visualize my resampled data (4096 Hz), all the channels look the same. It's not the case in my original data (16384 Hz). The data are sEEG data in .ncs file (neuralynx recording).
I attach some screenshots showing the way I do it. Is there any problem?
How can I solve this issue?
Thank you in adavance for your help!
Best,
Anaïs
1 Like
It could look like there are some differences in the reference or in the pre-processing.
Did you apply any montage (https://neuroimage.usc.edu/brainstorm/Tutorials/MontageEditor) or ICA/SSP cleaning procedure before resampling?
Make sure that there are no projectors applied to the data before resampling (Tab Record, menu Artifacts > Select active projectors), and that the same reference/montage is used in all the windows.
If there is something wrong with Brainstorm, I will need more information and maybe a short segment of data for debugging it.
Please post a screen capture of your figure (where the signals are all the same) on which you apply an average reference (Montage menu), in order to make sure that they are indeed all the same.
Thank you for your answer. Unfortunately, I did not find a solution...
The "Select active projectors" is empty.
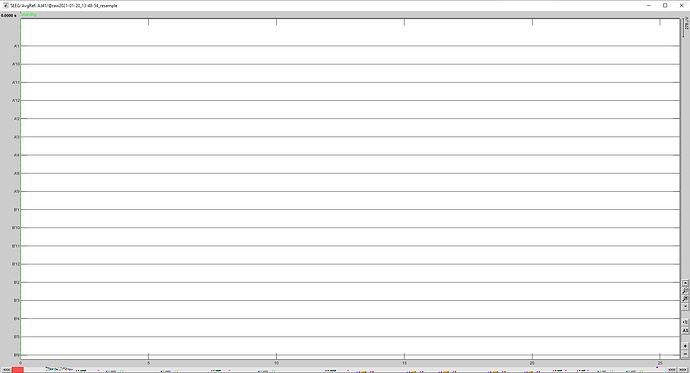
Here is a screen capture of my figure (where the signals are all the same) on which I have applied an average reference (Montage menu) in the Record tab. The signal is flat.
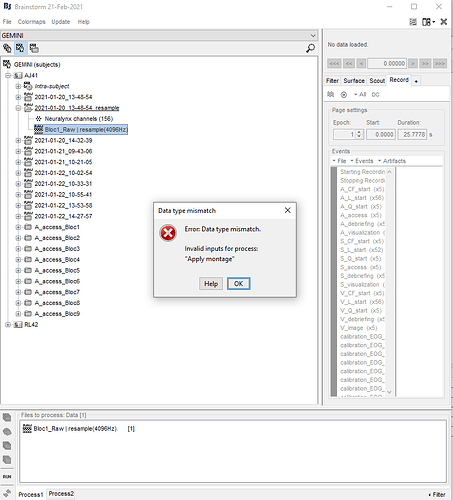
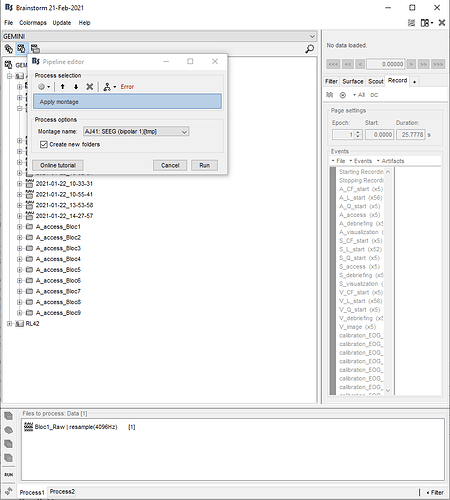
PS: I noticed another error. The menu "Apply montage" is not available and I get this error. It will be annoying for the next steps because I was planning to apply a bipolar montage. Could you help for this too?
One generic note before I keep going with the debugging: since you are resampling your data, you could maybe downsample it a bit more, eg. down to 2048 or even 1024Hz, unless you are really interested in trying to study frequencies above 300Hz.
Based on what you reported, there is probably something wrong in Brainstorm.
Could you please do the following:
- From your original data (16384Hz), right-click on the link > Import MEG/EEG, import a block of 1s (without using events).
- Try to resample this imported block
- Report here what the result is
- Right-click on the ORIGINAL 1s block > Review as raw
- Try to resample this new "raw" (=continuous) file
- Report the results here
- Right-click on the raw file created at step 4 > File > Show in file explorer
- Zip the content of the folder containing the file (should start with "@raw" and contain a file with a .bst extension)
- Upload this file somewhere and post the download link here (or send it to me as a private message on this forum)
The menu "Apply montage" is not available and I get this error.
For learning how to do this, please start by following the SEEG tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity
Thank you very much for your help!
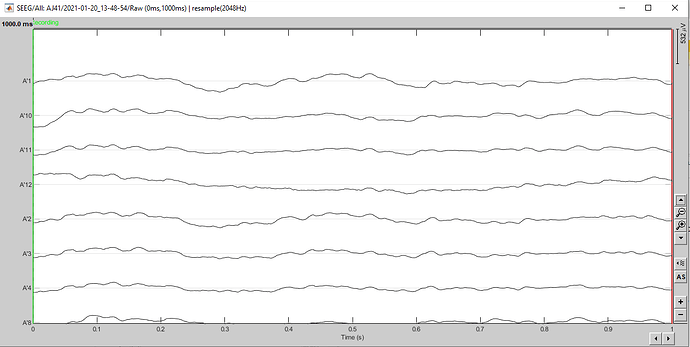
I have imported a block of 1s and I have resampled it (2048 Hz). Here is the result:
After, I review this 1s signal as raw. I resampled it. Here is the result:
Here is the link where you can find the file you have asked: http://depotcerco.univ-tlse3.fr/pupiph
I really hope that we can debug this. Thank you again for your help!
Thank you for the example file.
What I wanted to check is whether you have a problem with the resampling function.
Please post a screen capture of the IMPORTED + REVIEWED + RESAMPLED file (step 6 of my previous list).
For steps #3 and #6 (+re-import), I obtain the same signals (if we ignore some edge effects due to the resampling procedure). Nothing like what you reported in your first post.
Note that it is normal if they can't be displayed together: they have different sampling rates.
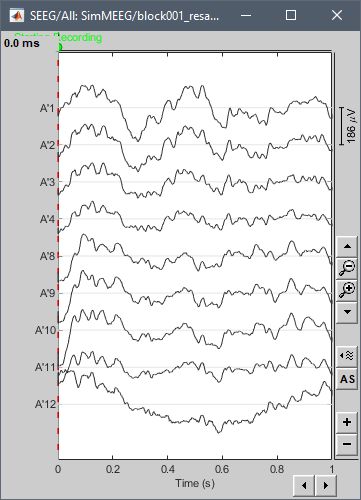
Hi François,
Here is the screen capture of the IMPORTED + REVIEWED + RESAMPLED file.
It's weird. Everything seems normal for me too when I use this 1s segment of the signal. But as soon as I try to resample all the signal, the bug appears again...
Unfortunately, if I don't manage to reproduce the error, it will difficult to fix it.
Do you have a short example file in the original format (neuralynx .ncs) that you could share?
(After making sure you do observe the same bug with it)
Do you observe the same issue if you run another process than the resampling?
(eg. low-pass filter)
I tried to do a low-pass filter. I had exactly the same problem. When I visualize the signal after this operation, all the channels are exactly the same.
Here is a link to a short example file. It is the data I used before except that I removed some channels. I did a test on it and I got the same problem than with the whole original file. http://depotcerco.univ-tlse3.fr/1vj2
Thank you for helping!
Thank you for the example file.
The error actually came from a bug in the Neuralynx reader, when reading channel by channel (which happens when applying frequency filters).
I think I fix it in this commit: https://github.com/brainstorm-tools/brainstorm3/commit/879cee1ae06feb56fce793234314a23e6154397a
Please update Brainstorm and try again.
It works now!
Thank you very much for your reactivity.