Hello,
I would like to do exactly the same kind of pipeline as Fran using a script to loop over subjects: computing different kernels and name them accordingy in the database (for example: constrained, unconstrained, etc... rather than having the date), and I was trying to rename the files using the process_add_tag after having called process_inverse_2018 :
%this is my script:
sFiles_constr = bst_process('CallProcess', 'process_inverse_2018', sFiles, [], ...
'output', 1, ... % Kernel only: shared
'inverse', struct(...
'Comment', 'MN: MEG ALL', ...
'InverseMethod', 'minnorm', ...
'InverseMeasure', 'amplitude', ...
'SourceOrient', {{'fixed'}}, ...
'Loose', 0.2, ...
'UseDepth', 1, ...
'WeightExp', 0.5, ...
'WeightLimit', 10, ...
'NoiseMethod', 'reg', ...
'NoiseReg', 0.1, ...
'SnrMethod', 'fixed', ...
'SnrRms', 1e-06, ...
'SnrFixed', 3, ...
'ComputeKernel', 1, ...
'DataTypes', {{'MEG GRAD', 'MEG MAG'}}));
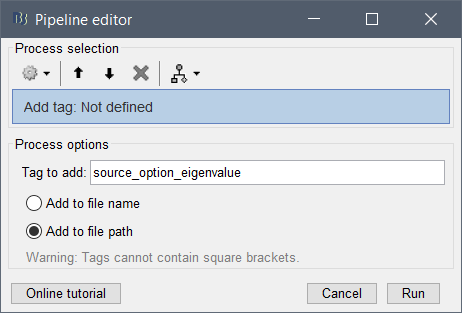
% Process: Add tag: _constr
bst_process('CallProcess', 'process_add_tag', sFiles_constr, [], ...
'tag', '_constr', ...
'output', 2); % Add to file path
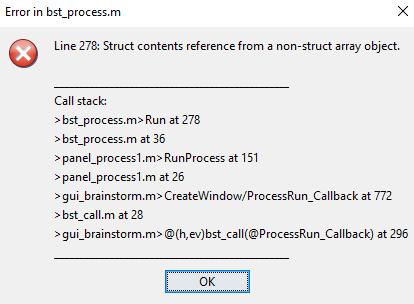
but I get following error:
Dot indexing is not supported for variables of this type.
Error in bst_process>Run (line 285)
allStudies = bst_get('Study', unique([sInputs.iStudy]));
Error in bst_process>CallProcess (line 2305)
OutputFiles = Run(sProcess, sInputs, sInputs2, 0);
Error in bst_process (line 38)
eval(macro_method);
Error in MYSCRIPT (line 48)
bst_process('CallProcess', 'process_add_tag', sFiles_constr, , ...
After what, the gui is seemingly unresponsive (turning wheel), and I cannot access File>View File Contents because I get this error:
Error using load
Unable to read file 'MYPATH/results_MN_MEG_GRAD_MEG_MAG_KERNEL_230427_0954.mat'. No such file or directory.
Error in view_struct (line 36)
MatContents = load(filename);
Error in tree_callbacks>@(h,ev)view_struct(filenameFull) (line 2403)
gui_component('MenuItem', jMenuFile, , 'View file contents', IconLoader.ICON_MATLAB, , @(h,ev)view_struct(filenameFull));
Which is indeed correct because the file HAS BEEN renamed in brainstorm_db with the tag _constr but somehow something is blocking the update...
Maybe relevant information: I am doing that at the Subject level and it appears under Common files because I have one headmodel per participant that I chose to share accross runs.
Thank you!
Julie