Hello Raymundo, thanks for this input ![]()
Sorry to keep bothering you but the matter is a bit more complicated and not resolved yet.
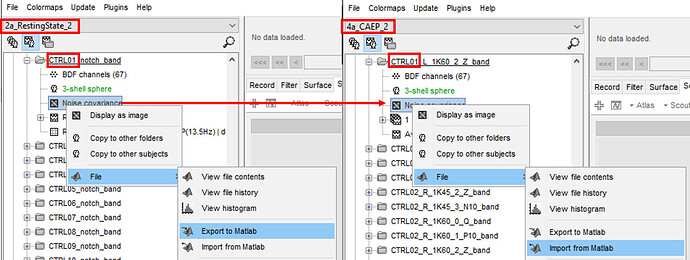
First, let me explain why the noise_cov was manipulated outside of Brainstorm. The noise_cov was computed from a different Brainstorm protocol which was a study on resting-state EEG. My cortical auditory evoked potential (CAEP) and resting-state EEG (RS-EEG) studies are in different protocols. I wanted to use the noise_cov computed from the RS-EEG protocol from the same participant and import that noise_cov into the same participant in the CAEP protocol. I chose not to use pre-stimulus baseline to compute noise_cov as I thought the duration of the pre-stimulus baseline was too short (200 m; with 1024 Hz sampling frequency). Whereas RS-EEG recording duration was 5 mins (with 1024 Hz sampling frequency). The below screenshot shows what I've done; noise_cov from CTRL01 participant from RS-EEG protocol was exported into Matlab workspace and then imported into the CTRL01 participant CAEP file. As there were multiple CAEP conditions (e.g. different stimulus intensities), the same noise_cov file from the same participant from the RS-EEG protocol was imported into multiple folders with the same participant.
Now comes the confusing part... As you rightfully pointed out, the imported noies_cov file did not have nSamples and I saw the same warning that you mentioned. However, a) the problem noise_cov did not have missing fields, and b) nSamples was absent even in folders where everything worked fine. This is odd because if the imported noise_cov file was the issue, all folders from the same participant should have the same issue but this problem is selective to certain folders. I checked to see if the imported noise_cov files were somehow different between folders from the same participant but there was no difference to the noise_cov data between folders and nSamples was missing in both the problematic folder and the correctly working folder. So I do not think the absence of nSamples was the issue as all other source computations managed to work well without nSamples.
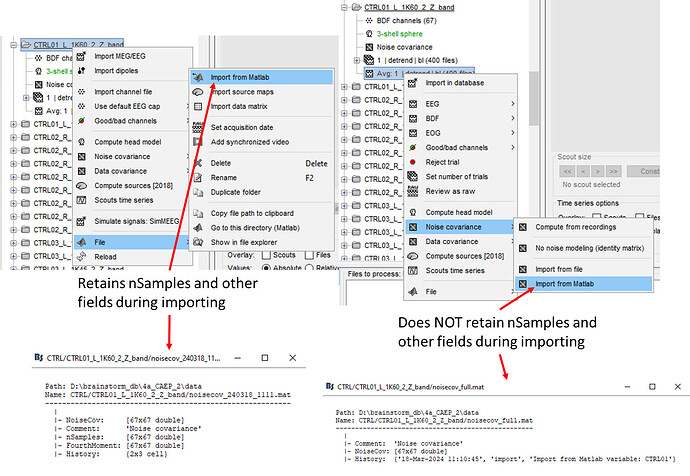
In attempting to troubleshoot this problem I re-exported the noise_cov file from CTRL01 from RS-EEG protocol into Matlab workspace possibly thinking that the exporting process had issues. Interestingly, the noise_cov file that I just exported did contain nSamples as one of the fields. It wasn't until later that I realised that the issue was during the IMPORT process. If you import the exported noise_cov file using folder click > file > import from Matlab, everything that you've exported (as in the entire struct containing nSamples and, etc) is imported. However, if you import the exported noise_cov file using averaged trial recording click > noise covariance > import from Matlab, then several fields including nSamples is removed from the struct during importing (noise_cov data is identical; just with missing fields from struct). Not sure why this is the case maybe the BST team may need to look into this?
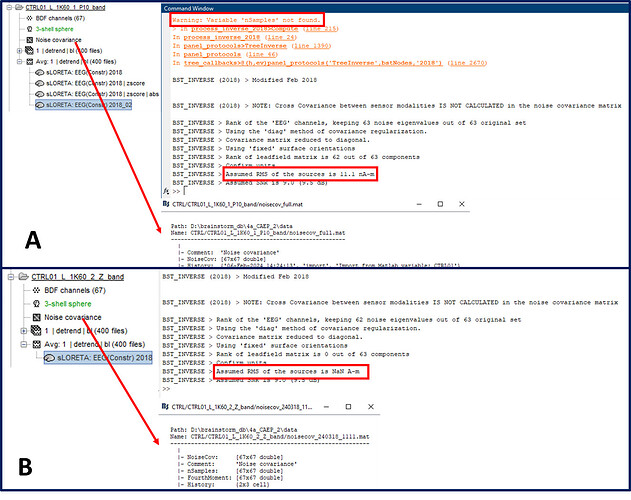
Anyway, subsequently to the above, I first deleted the noise_cov that was the issue and re-imported this new noise_cov into the folder with issues (CTRL01_L_1K60_2_Z_band) and ran compute sources. As expected, due to the presence of nSamples there were no warning messages. However, the source computation values were all still showing NaN. So I compared the messages that Bst gave me in the Matlab command window when I ran compute sources [2018] between the good and the bad folders and the following was what I observed. A and B shows the outputs from the "good" and "bad" folders respectively. A: has noise_cov that does not have nSamples and other fields due to being imported through "averaged trial recording click > noise covariance > import from Matlab" and therefore you can see in the Matlab command window "Warning: Variable nSamples not found". B: has noise_cov that contains nSamples and other fields due to being imported through "folder click > file > import from Matlab" and hence you cannot see the same warning (I also show the noise_cov file contents to further illustrate my point). I should remind you that he noise_cov data itself are identical between folders.
What is troubling is the part that says "Assumed RMS of..." I think this is the part that is causing the issues. A, which does not have nSamples is still showing a numerical value for "Assumed RMS of..." but B even though it has all the fields of noise_cov shows "Assumed RMS of the sources is NaN A-m". Although I think this is the issue I have no clue how to correct for this.
What do you think about all this? I share the folders that I am showing in the figure immediately above. I've labelled them GOOD and BAD to illustrate which is which. My apologies for such a lengthy and detailed post (just want to properly troubleshoot this phenomenon with your help). Many thanks for your time in advance.
CTRL01_L_1K60_1_P10_band_GOOD.zip
CTRL01_L_1K60_2_Z_band_BAD.zip