Hi, I met a problem about the compiled version of Brainstorm during importing the raw datarecord in Compumedics ProFusion Sleep .rda format.
Following the official tutorials I developed a script to do the task. It runs well in my local Matlab environment with correct full data importing and exporting to SPM data.
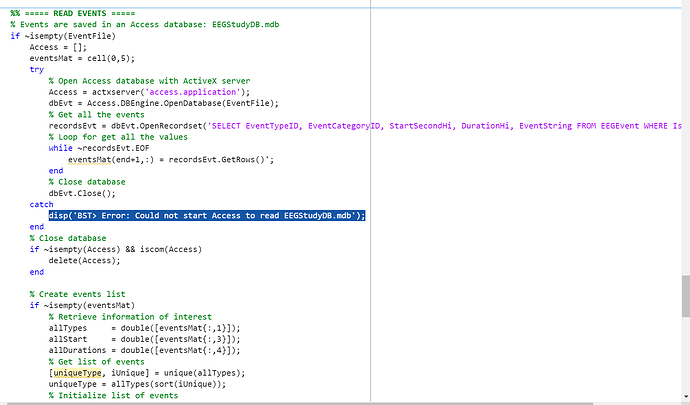
However when I deploy it into cluster with correct Matlab Runtime and officially compiled Brainstorm installed, it occurs that the EEGStudyDB.mdb file cannot be accessed which means the annotations are not imported successfully.
I checked the source codes in in_fopen_compumedics_pfs.m to see the block that cause the error message. I suppose the activeX may be the reason why this script perform well in local full installation of MATLAB but failed in Runtime.
Can you be helpful for this problem? Thank you so much!
ErrorMessage:
BST> Starting Brainstorm:
BST> =================================
BST> BrainstormHomeDir = /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/bst_javabuil
BST> Version: 19-Apr-2024
BST> Deleting old process reports...
BST> Loading configuration file...
BST> Checking internet connectivity... ok
BST> Plugin brain2mesh: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/brain2mesh
BST> Plugin iso2mesh: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/iso2mesh
BST> Plugin ct2mrireg: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/ct2mrireg
BST> Plugin brainentropy: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/brainentropy
BST> Plugin blackrock: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/blackrock
BST> Plugin easyh5: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/easyh5
BST> Plugin neuroelectrics: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/neuroelectrics
BST> Plugin jsnirfy: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/jsnirfy
BST> Plugin xdf: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/xdf
BST> Plugin libsvm: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/libsvm
BST> Plugin fastica: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/fastica
BST> Plugin picard: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/picard
BST> Plugin derivelfp: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/derivelfp
BST> Plugin nirstorm: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/nirstorm
BST> Plugin mcxlab-cl: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/mcxlab-cl
BST> Plugin mia: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/plugins/mia
BST> Plugin spm12: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/spmtrip
BST> Reading process folder...
BST> Database folder: /mnt/gluster/home/chencheng/.brainstorm/local_db
BST> Loading current protocol...
BST> =================================
WARNING: This file is a function, trying to convert to a script...
BST> Error: Could not start Access to read EEGStudyDB.mdb
BST> Checking latest online version for spm12...
BST> Plugin spm12 already loaded: /mnt/data/gv0/home/chencheng/.mcrCache9.13/bst_ja0/Users/rcassani/.brainstorm/spmtrip
BST: File "Subject01/@rawRDA-6/data_0raw_EEGData-6.mat" exported as "/mnt/data/gv0/home/chencheng/HongaiRDA/Exported_HongaiRDA/EEGData-6.mat"
BST> Brainstorm stopped.
BST> Compiled code returned without errors.