Hello Brainstorm Team,
I am trying extract the scout time series in MATLAB in order to run coherence. I have a lot of subjects (178) so I need to automate this process. In trying to automate this I have noticed I can either run extract scout time series and proceed to coherence brainstorm script or I can apply the scouts in the coherence script. However, whenever I do either the atlas I have chosen is not transferred over.
In my code I am pulling in the correct file (the same mat file that I pull down in the gui) as the sfile:
sFiles = wfu_find_files('data_0raw',session_dir);
sFiles = char(sFiles);
% Start a new report
bst_report('Start', sFiles);
Next I run this code:
% Process: Scouts time series: [68 scouts]
sFiles = bst_process('CallProcess', 'process_extract_scout', sFiles, [], ...
'timewindow', [0, 4], ...
'scouts', {'Desikan-Killiany', {'bankssts L', 'bankssts R', 'caudalanteriorcingulate L', 'caudalanteriorcingulate R', 'caudalmiddlefrontal L', 'caudalmiddlefrontal R', 'cuneus L', 'cuneus R', 'entorhinal L', 'entorhinal R', 'frontalpole L', 'frontalpole R', 'fusiform L', 'fusiform R', 'inferiorparietal L', 'inferiorparietal R', 'inferiortemporal L', 'inferiortemporal R', 'insula L', 'insula R', 'isthmuscingulate L', 'isthmuscingulate R', 'lateraloccipital L', 'lateraloccipital R', 'lateralorbitofrontal L', 'lateralorbitofrontal R', 'lingual L', 'lingual R', 'medialorbitofrontal L', 'medialorbitofrontal R', 'middletemporal L', 'middletemporal R', 'paracentral L', 'paracentral R', 'parahippocampal L', 'parahippocampal R', 'parsopercularis L', 'parsopercularis R', 'parsorbitalis L', 'parsorbitalis R', 'parstriangularis L', 'parstriangularis R', 'pericalcarine L', 'pericalcarine R', 'postcentral L', 'postcentral R', 'posteriorcingulate L', 'posteriorcingulate R', 'precentral L', 'precentral R', 'precuneus L', 'precuneus R', 'rostralanteriorcingulate L', 'rostralanteriorcingulate R', 'rostralmiddlefrontal L', 'rostralmiddlefrontal R', 'superiorfrontal L', 'superiorfrontal R', 'superiorparietal L', 'superiorparietal R', 'superiortemporal L', 'superiortemporal R', 'supramarginal L', 'supramarginal R', 'temporalpole L', 'temporalpole R', 'transversetemporal L', 'transversetemporal R'}}, ...
'scoutfunc', 3, ... % PCA
'isflip', 1, ...
'isnorm', 0, ...
'concatenate', 0, ...
'save', 1, ...
'addrowcomment', 1, ...
'addfilecomment', 1);
This results in the sFiles being cleared out entirely from the MATLAB workspace.
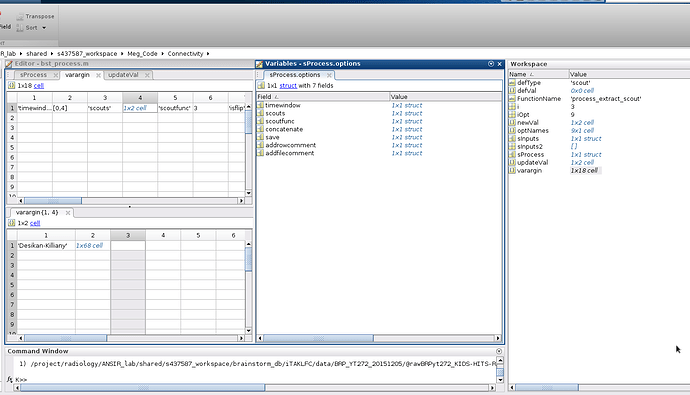
After looking closer at the issue in the bst_process.m file line 2130 results in answers =1 but in line 2131 the defVal is .
I can only post one picture so here is what everything looks like during the for loop in the bst_process after running the first i which is "timewindow"...
Could you please help me with this issue I am running into ?