Hi @Francois,
I am still working on my analysis in BST (Nirstorm). I managed to solve the resample problems by following your instructions. Thanks!
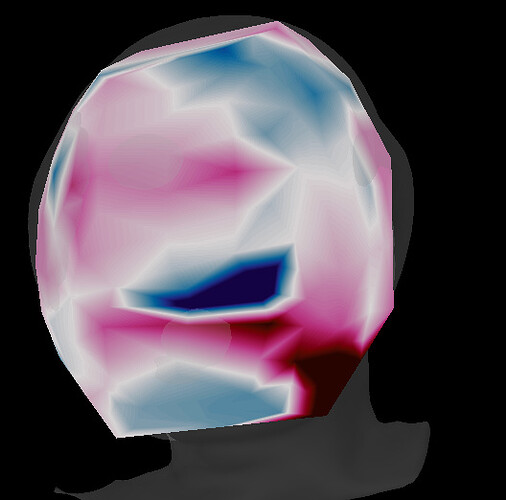
Now I am facing another issue. When I compute a big average of the trials (using the averages of the trials of each subject) I have some problem in 3D sensor cap. This happened in two of the four groups in my analysis, and I made the same previous steps for all groups.
I don’t know if it is a relevant point, but when I compute Power test against the baseline using the same files, there is no problem with the sensor cap image at all. When I compute Permutation test using at least one “problem group”, the sensor cap image is also disrupted.
Do you know how can I solve it, please?
Thank you for all your help!!
A good one:
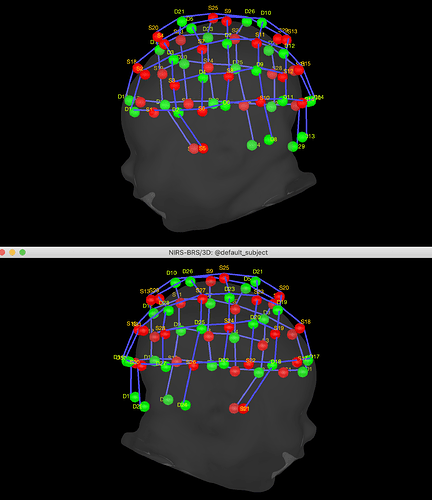
I guess the problem is not related with averaging the channel files, but with averaging the 3D positions of sources and detectors. Do you have exactly the same list of channels in the channel files of all the subjects? (same number, same order?)
If there is no channel file in the output folder, then Brainstorm averages the locations in the input channel files. This might not work well with NIRS...
For further debugging:
- Looking for two subjects that when averaged, give a result you don't like
- Post a screen captures of the sensors (double-click on the channel file)
- Zip together the channel file of the two subjects and one data file in each, upload it somewhere and post the download link here.
@tvincent @edelaire
Are you supposed to average fNIRS data like this?
Should we add some extra checks or average the fNIRS channel files in a different way?
We have a channel file for every subject, with same number and order.
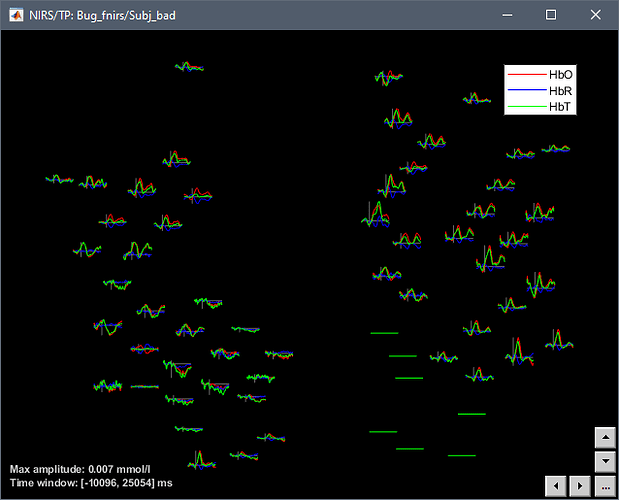
You can find good and bad subject here: Dropbox - File Deleted - Simplify your life
Follow below: error message and screen capture
Thanks!
Error using matlab.graphics.axis.Axes/set
Error setting property 'CLim' of class 'Axes':
Value must be a 1x2 vector of numeric type in which the second element is larger than the first and may be Inf
Error in figure_topo>UpdateTopoPlot (line 160)
set(hAxes, 'CLim', CLim);
Error in figure_topo>PlotFigure (line 651)
UpdateTopoPlot(iDS, iFig);
Error in figure_topo (line 27)
eval(macro_method);
Error in view_topography (line 385)
isOk = figure_topo('PlotFigure', iDS, iFig, 1);
Error in tree_callbacks>@(h,ev)view_topography(FileName,Modality,'3DSensorCap') (line 2817)
gui_component('MenuItem', jMenu, [], '3D Sensor cap', IconLoader.ICON_TOPOGRAPHY, [], @(h,ev)view_topography(FileName, Modality, '3DSensorCap'));
Thanks for the example data.
The problem is that the files include some NaN values.
I added a check in the topo plot functions so that it replaces the NaN values with zeros: Bugfix: Topo plots: Removing NaN from data · brainstorm-tools/brainstorm3@6a5548c · GitHub
Update Brainstorm to get this fix.
However, this is only a patch on a more general problem:
How did these NaN values ended up here? I guess it means that you have no data for these sensors, but this not the way bad channels should be handled in Brainstorm. If there is no data, the values should be ZEROS (not NaN) and the corresponding values in ChannelFlag should be set to -1.
See reference doc: https://neuroimage.usc.edu/brainstorm/Tutorials/Epoching#On_the_hard_drive
@tvincent @edelaire
Should this be fixed somewhere in your reading functions?
It worked! Now we have beautiful 3D-sensor-caps in our data! Thank you!
The topo plots work but many other computation functions (averaging, statistics...) may have problems because of these NaN values, you should try to fix them, and in all cases: at least mark all these channels as bad.