Hello All
Im trying generate simulate sources with the interface of BST.
I have mi head model ( BEM model).
I know create simulations from the simulated signal (generic signal) and one scout.
I need use 18 differents scouts and use the same simulated signal in each scout (18).
Is possible perform this? how?
I tried create two different simulations signal from two differents scouts, and BST show me a error.
The steps that I do are the followings:
- create a generic signal
- move the simulated signal to “Process 1” and “run”
- simulate recording from scouts
( Inside this process I tried choose ( for test) two diferent scout for two differents simulations, and show me error).
I hope you can help me, because is too much work perfom the simulation one by one.
Regards
What error are you getting?
Hi all
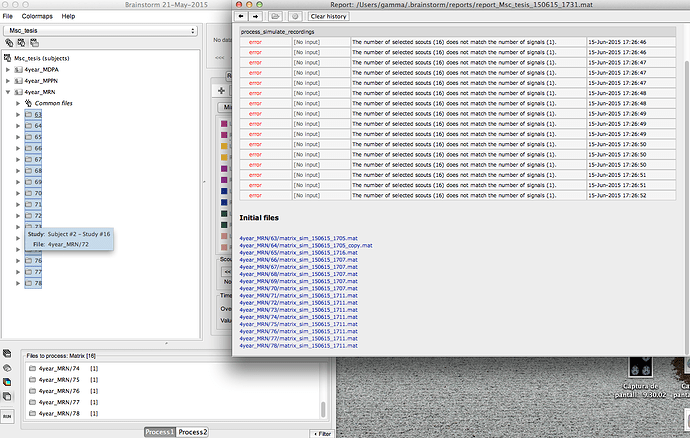
I have the next error
I want to generate 16 simulations from 16 differents scouts, with the same generic signal.
And my error is showed in the image
Thanks for you help
Hello,
If you use 16 scouts in input, you need to select a file with 16 scouts time series. This is why you get this error.
However, I’m not sure this is what you want to do. This would not loop over the 16 scouts but simulate recordings when all the 16 scouts are active at the same time.
If you want to automate this: write your own scripts.
In the pipeline editor window, instead of clicking on “Run” to execute the selected process, use the menu “Generate .m script”.
Then edit the script to add your loops, and all the logic you need to manage your simulations.
Cheers,
Francois
Thanks Francois
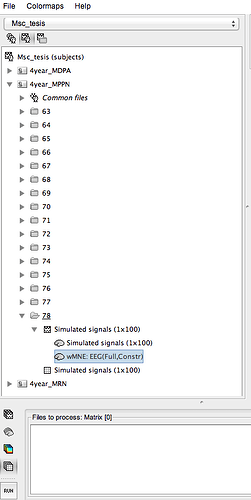
I could do what i wanted, however i wanted open the file wMNE: EEG(Full,Constr) through command window. To after add to line code in my function.
Can you say me how do this?
the file you wish to open for command line shown in the picture
Regards
Hello,
To open a 3D view of source results, you can use the function: view_surface_data([], SourceFile)
Many other examples are available in tutorial script: brainstorm3/toolbox/script/tutorial_ctf.m
Francois
Thank you for your response francois
I do not need wMNE display the file. However I need to just export the file from Matlab code line, then I could change the wMNE file. I know I can export the file BST interface to Matlab, but I need to export it from line of code, and this is what I need to learn.
Greetings and sorry for the insistence.
This is a Matlab .mat file.
You can load it from your script and with Matlab’s load() function, and save it back with the save() function.
Francois sorry for my insistence
I can not yet do what i need..
For clearing my question I pasted a image with the code
I have a code as this:
Watch the line 57. I need save the generated file wMNE: EEG(Full, Constr) in every loop of my cycle (for)
whenever the resolution of the inverse problem wMNE run, save this in a variable as wMNE. Because I need to work with this variable
Regards
You need to use the path of the file you want to load, not its comment.
ResultFile = file_fullpath(sFiles(1).FileName);
ResultsMat = load(ResultFile);
FYI: To address all the recent questions we had related with simulations, I added some code in Brainstorm and wrote a new tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/Simulations