Hello,
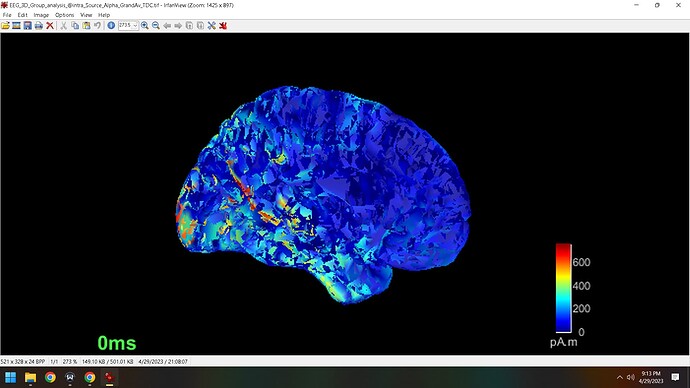
I was attempting to overlay a source map on a mask without sources and to my surprise the medial view image appears to now show the dataset in polygons, which looks very messy. I tried to reset to default settings to return back to its original image but I am stuck with this new representation of the source maps. Can you help me understand what I must do to revert back to the original setting of the source map?
Best,
Zach
Can you more clearly explain what you did? If you added twice the same surface, the faces would be overlaid and probably look strange like this. You can see the surfaces you have loaded at the top of the surfaces panel:
Where you could remove the extra surface. But also just closing the figure and reopening the result should clear up any added surfaces.
Or if you did some computation directly in Matlab, maybe you ended up with NaNs? You can also just rerun the inverse model to get a fresh source map.
Hello,
I have to return to this post becuase the issue was never solved. I attempted to determine if there were two surfaces uploaded but that doesn't seem to be the case. Opening and closing does not help. This affects all slices, but can only be seen visually on medial slices, and can be seen in all slices when altering the level of 'transparency' in the surface options tab. I did not run anything directly in Matlab.
I have tried uploading the version of Brainstorm, restarting Brainstorm and Matlab. I'm not sure if I accidently selected an option that caused this to change but if so, perhaps it has something to do withthe number of vertices which no longer match. Any ideas?
Zach
If it helps, its showing that the number of vertices is 15002 and the number of faces is 29984
It may be the case that the sources were computed with one surface file, and then the surface was modified.
Can you describe what were the steps you made?
- How was the surface obtained?
- How were the sources computed?
Does the surface related to the sources seem correct?
- Right-click on the source file (
 or
or  ) > File > View contents and check the
) > File > View contents and check the SufaceFile field
- Go to the Anatomy view and display that Surface file (hover on the files to verify it's the same file)
- You can activate the Edge option int he Surface tab.
- Do no use transparency, as the vertices (and edges) from the lateral side will be also shown and the it will be difficult to see the figure.
Post images of the Surface, and the Sources using the save view with 0% transparency. You can press the key [=] in one figure, so its view will be used of all the open figures.
I computed sources using the following features:
Kernel Only: One per file
Minimum Norm Imaging
sLORETA
Constrained
A noise covariance calculation was made prior to, along with a head model using OpenMEEG Boundary element model with a 15002x3 grid.
In the viewer contents page, it says @default_subject/tess_cortex_pial_low.mat which matches the anatomical view with 15002 vertices.
test1
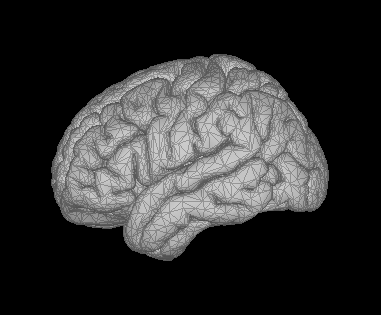
test2
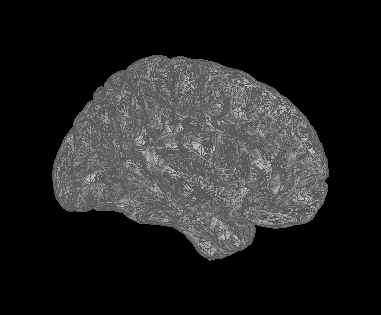
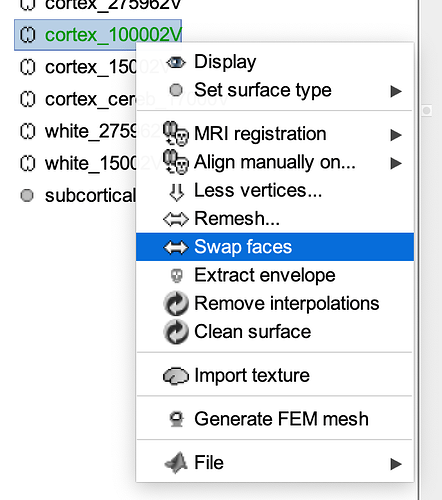
You may want to try the "swap faces" option that is available when you right-click over the cortical surface you use for deriving source maps. You can do that from the anatomy data tree, then recompute the head model and source map.
1 Like
Test1 and Test2 are arbitrary names. They are lateral and medial slices. The issue occurs for medial slices with 0% transparency and but also for the lateral slices with some degree of transparency.
I'm running an analysis at the moment. After I will attempt the 'sap faces' route
I see. Can you display only the medial view of that hemisphere but without any source map, just the anatomy, please? It could be that the medial view display cuts though the medial wall of the hemisphere. You can also add a full view of the two hemisphere together, say from the top and and side, with the sensors added on top, please.
If possible upload images with better resolution.
Also, images (.bmp, .jpeg and .png) can be drag-and-drop in the comment box
The test2 image is a medial view of the anatomy.
Swap facs has not recovered the image I need. Recalc sources at this point is quite ardous from here as it took a lot of time and I have found a way to display medial view with this issue arising by selecting the Struct option in data options. The problem seems to go away using this option. With full view there is no issue unless I make the image transparent to some degree.
Thanks.
So I think what's happening is due to the fact that the left/right hemisphere are not properly identified by default by Brainstorm. This means one of the axes of the coordinate system is not properly aligned along the sagittal direction between the two hemispheres. This is fine, as long as you make sure the head sensors are properly aligned with respect to the anatomy in that coordinate system , hence my recommendation to add the sensor locations to these 3D views.