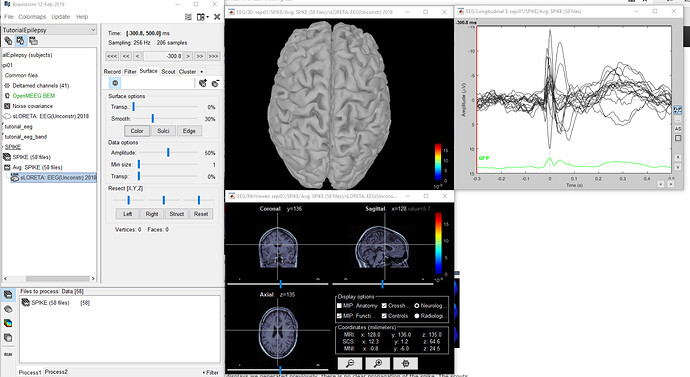
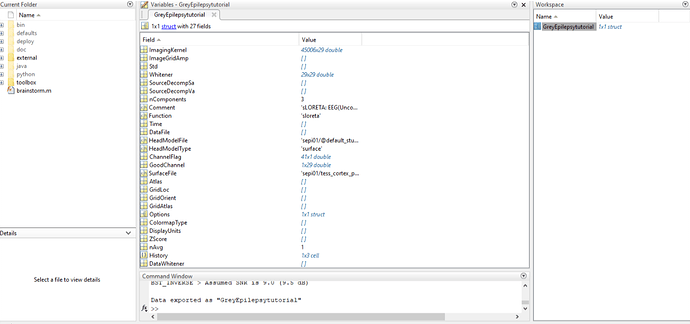
I have been unable to get a cortex model when computing sources for my personal data. The patient data shows a seizure which I am looking to localize, however all attempts to compute sources present a blank/grey brain or only the MRI view, which is also blank. I have followed a number of tutorials for computing the head model, noise covariance, computing sources, volume sources, deep brain analysis... all gave me a grey brain. I finally decided to check brainstorm by downloading the epilepsy data from the brainstorm download page and following the tutorial EEG and Epilepsy exactly. I had all of the same problems: the cortex is grey; the MRI viewer shows nothing; when imported into MATLAB, the Imaging Kernel contains data but the visual matrices are empty.
I am not seeing this issue on the forum so I am guessing that there is an issue with my installation. What could cause this? Why am I only seeing this issue with source localization but the rest of the program appears to work fine? Am I correct that this is corrupted or improperly installed?
I will attempt to reinstall and try again, then give an update.
Below are pictures of my results and the file in MATLAB.
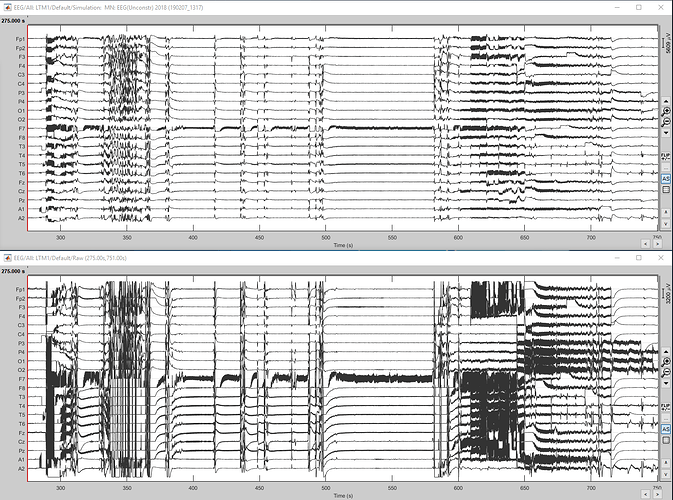
In a previous attempt with my own data I ran a current density source computation and ran Model Evaluation > Simulate recordings. The results are below, the top is the simulated and the bottom is the original. That is how I could tell that the data for the source localization was present in some capacity, even if I could not get a visual.
Maybe it is a matter of scaling the colormap to your data. For this, right click over the colorbar in the 3D brain figure and select from the colormap submenu and select 'Maximum: Local'.
From the main GUI panel, you can also bring the Data Options: Amplitude threshold down to 0 to look at the entire brain map.
You also need to move your time cursor to the time of interest (0s for the epilepsy dataset)
Thank you, I finally see activation in my seizure patient. The activation does not change when I move the cursor though, so it is hard to tell where it is located. Is there a way to fix this?
What time cursor are you referring to? Also, I can only view the MRI (3D), and MRI (MRI viewer), but not the cortex activations. Is this normal?
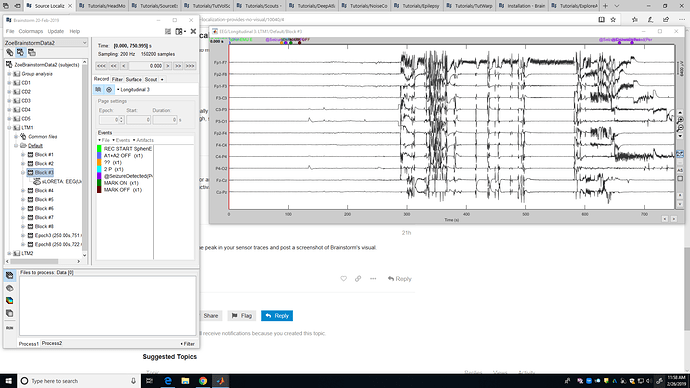
Please go at one peak in your sensor traces and post a screenshot of Brainstorm's visual.
Thanks.
Is this what you are asking for? This is a block of my EEG data in which the subject was having a seizure.
I recommend you start by reading the introduction tutorials (at least until #23) using the example datasets before trying to process your own recordings. I guess most of your questions will be addressed in there.
https://neuroimage.usc.edu/brainstorm/Tutorials
For processing EEG recordings, I recommend you also follow the EEG/Epilepsy tutorial:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy
I have read almost every tutorial 1-23 and, as I stated in the original post, "I finally decided to check brainstorm by downloading the epilepsy data from the brainstorm download page and following the tutorial EEG and Epilepsy exactly" with the tutorial dataset. The terminology used in your requests is unfamiliar to me even after reading the tutorials and searching the brainstorm site for such a phrase. If there is a tutorial explaining the connection between the time cursor and the source localization or what a sensor trace is, please direct me to it and I will read it.
Time cursor:
https://neuroimage.usc.edu/brainstorm/Tutorials/ReviewRaw?highlight=(time+cursor)#Navigate_in_time
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy?highlight=(time+cursor)#Manual_marking
"Connection between the time cursor and the source localization":
https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation?highlight=(Change+the+current+time)#Display:_Cortex_surface
Sensor trace = EEG signals = EEG time series
I can not believe I missed that. Moving the time cursor changes the visual and gives me the activation I have been looking for. I will run the analysis again to see if I can get the cortical activations rather than just the MRI viewers. Thank you.