Hi,
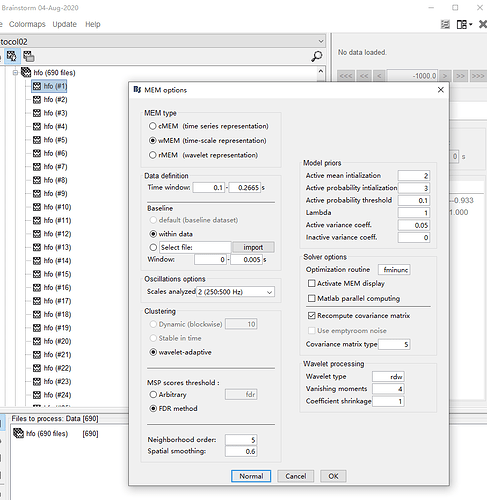
I'm trying to compute source localization of high frequency oscillations with wMEM. But errors occur:
The index exceeds the matrix dimension.
Error process_inverse_2018>Compute (line 667)
OPTIONS.Data = DataMat.F(GoodChannel,:);
Error process_inverse_2018 (line 24)
eval(macro_method);
Error panel_protocols>TreeInverse (line 1316)
[OutputFiles, errMessage] = process_inverse_2018('Compute', iStudies, iDatas);
Error panel_protocols (line 44)
eval(macro_method);
Error tree_callbacks>@(h,ev)panel_protocols('TreeInverse',bstNodes,'2018') (line 2528)
gui_component('MenuItem', jPopup, , 'Compute sources [2018]',
IconLoader.ICON_RESULTS, , @(h,ev)panel_protocols('TreeInverse', bstNodes, '2018'));
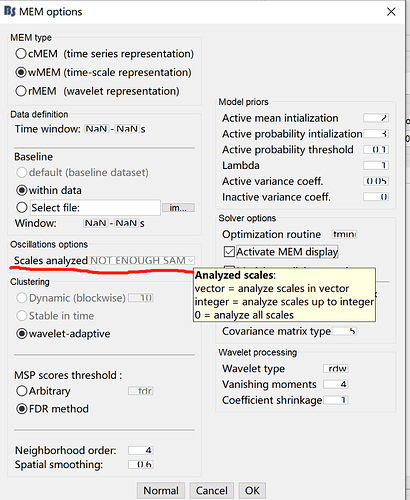
Could you help me solve the problem? And what does it mean by "Not enough in SAM" when set "Oscillations Options"? Thanks