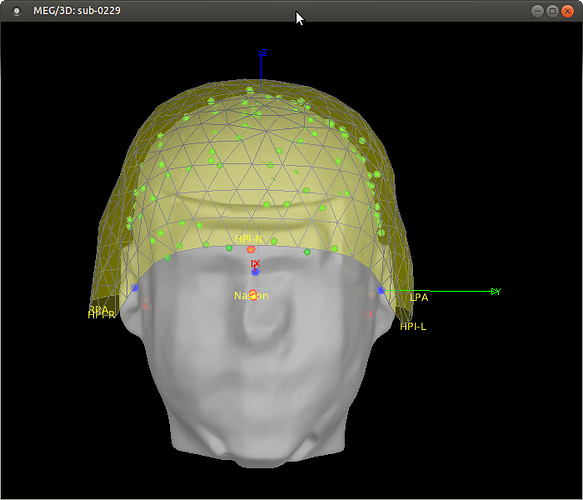
I was a little confused about the subject coordinate system after using "refine with head points". See the attached screenshot. It appears the SCS axes stay aligned with the anatomy (MRI) whereas I expected them to follow the head points, such that for example the X axis would still go through the nasion point from the digitization, and not the nasion picked on the MRI. I thought the advantage of digitizing the anatomical locations was that it would be more precise than manually picking them on the MRI. But it seems Brainstorm is designed with the opposite viewpoint, keeping the MRI fiducials and ignoring the digitized anatomical locations after refining the co-registration. I don't have a specific question, but I'm curious to hear your thoughts or comments about this choice.
The spatial reference on which everything is aligned is the SCS referential based on the MRI anatomical landmarks. These points are defined once at the level of the anatomy and may not change across multiple experiments.
When importing MEG/EEG/NIRS sensor positions, these are converted to the referential defined by these anatomical landmarks: a rigid transformation is applied to the positions of all the points residing in the sensor space (electrodes, MEG coils, optodes, head localization coils, other headpoints). Anatomical landmarks always remain unchanged.
As suggested, I'm copying another related email thread here.
Hi Mark,
The function channel_detect_type has an isAlign option, but it uses the digitized anatomical points from the channel file to do the alignment, not those from the MRI. This seems to conflict with what is done elsewhere. (As I had observed a few years back.)
And my reply to this comment would be exactly the same as three years ago.
When warping the default anatomy to head points, from what I can tell the coordinate system is not changed. bst_warp has a section for warping fiducials, but the 3 points that define SCS are not warped (commented out). So the NAS,LPA,RPA points no longer match the now warped anatomical points. I agree that for a blurry template that's not very important, and it saves us having to update everything. But it does make the error greater if some transformations are done with digitized points as above and others with MRI points.
I'm not sure I understand this point: After warping, you're not supposed to adjust the MRI/sensors registration anymore.
If you think there is anything wrong in the current warping workflow, could you please assemble a clear example I can reproduce on my end?
Please post your messages in the old thread you mentioned before, so we these conversations are archived in a structured way.
Finally, for head tracking, it uses both warping to get a scalp surface, and channel_detect_type to align. So I think I have to fix that, but I wanted to double check with you first if channel_detect_type should also be changed.
The head tracking code may have issues related with a wrong sequence of registration calls. You may need to debug this, indeed.
The function channel_detect_type is used intensively by all Brainstorm users. Changes to it would be complicated to test, and may alter hundreds of existing processing pipelines. I would prefer avoiding any modification to it unless there is something really wrong with it.
The head tracking is used at most by a handful of users, including you.
If anything needs to be modified for fixing the head tracking, I'd suggest it is done on the head tracking side only, even if it's a bit hacky.
Does it make sense?
Francois
Hi François,
Please have a closer look at my email, especially the first point. I think there is something wrong with channel_detect_type using digitized anatomical points. Your previous answer to this forum thread basically just confirmed that Brainstorm uses MRI anatomical points to define the SCS coordinates. So the MRI ones should be used everywhere and the digitized ones not used at all since they are different and basically "arbitrary" (but following a convention). Here I'm pointing out an inconsistency where the sensors and anatomy would be using coordinate systems based on different points.
The point about warping is that since the MRI NAS etc are not warped, they will definitely not match the digitized anatomical points. So if the latter are used to align sensor coordinates (as in channel_detect_type), then the misalignment is greater. This would be fixed if everything uses the MRI points.
My point about head tracking is secondary, I just want to resolve this first and then I can adjust it accordingly.
Thanks!
Marc
Of course, they have to.
- In the MRI, we mark manually the points on the reference MRI volume using the MRI viewer (or using some automatic processing).
- In the sensor space, we get the points from a 3D digitizer or from the MEG (localization of the head tracking coils) - With the specific sub-case where we do both, and correct the MEG sensors based on the detected coils to use digitized anatomical landmarks
- The two things are done independently, and the combined result of the two procedures is our coregistration sensors/MRI. Using the MRI landmarks in the sensor space (eg. in channel_detect_type) is not possible, as we initially don't know how to transfer MRI coordinates in the same space as the sensors.
I'm really sorry if I'm still missing your point. It looks like there is something obviously wrong that you're telling me to look at, but I can't see it, I'm probably not looking at the place or along the right angle. Maybe having the full workflow laid out would help me understand.
Can you please assemble an full example illustrating your point?
- starting from the import of the MRI and the import of the first MEG run of the tutorial dataset,
- following the steps of the introduction tutorial,
- illustrating step-by-step what happens with the various coordinate systems (R and T matrices) and fiducials coordinates (in the MRI and in the channel file).
- when the debugger hits
channel_detect_type.m: explain why the code is wrong, where exactly, and what would you replace it with
The point about warping is that since the MRI NAS etc are not warped, they will definitely not match the digitized anatomical points. So if the latter are used to align sensor coordinates (as in channel_detect_type), then the misalignment is greater.
Is your issue specific about importing new MEG data AFTER warping the anatomy?
This is indeed not the proposed workflow, which is to 1) import all the MEG data, 2) warp the anatomy, 3) never modify the registration of the sensors again.
Could you please write a full sequence of operations that illustrates this issue, based again on the example dataset (maybe importing the second MEG run after warping the anatomy with the first one), so that we can clearly identify at which stage the issue is? And what action we need to take?
I guess it could take two directions: when importing data in a subject with a warped anatomy, we could either call channel_detect_type.m in a different way (or completely skip it and do something else), or giving a warning to the user saying that all the data should be imported BEFORE warping the anatomy.
This would be fixed if everything uses the MRI points.
How do you use the MRI points to align the sensors?
That would result in computing the SCS transformation (R+T) based on the results of the previously computed SCS transformation, without which we can't transfer the MRI fiducials into the space of the sensors...
Thanks for your patience.
Not at all, than you François for helping me understand.
I have thought about it more in detail like you suggested and will try a few examples later. I think you're right that the usual workflows are ok.
- MEG sensors are aligned with head points through the 3 digitized head coils.
- These are all (temporarily) converted to a coordinate system based on the digitized anatomical landmarks.
- Anatomy files (MRI, surfaces, etc) are converted to a coordinate system based on anatomical landmarks placed on the MRI.
At this point, there is a mismatch, based on how well the 2 sets of landmarks (digitized vs MRI) match. (This is one thing that was wrongly concerning me, because I had in mind that the "refine" or warping process had already happened. In practice, we wouldn't have to go to the digitized anatomical coordinates if we're going to use the "refine" process.)
- Then, the head points are aligned with the MRI anatomy with the "refine" process. This converts sensors and head points to the MRI-based coordinate system and everything is ok.
or
- Warping a template. It was not as clear how this works, but from the tutorial, I think the logic is that we first do manual registration where we match the 2 coordinate systems at step 3. Then warping displaces as well as deforms the MRI to match the head points, so that everything then really ends up in the digitized anatomical fiducials coordinate system. So while both sets of fiducials won't be exactly the same, they should both define the same coordinate system axes.
If this is all correct, I can now return to the head tracking code with a better understanding.
Thanks!
Marc
Then, the head points are aligned with the MRI anatomy with the "refine" process. This converts sensors and head points to the MRI-based coordinate system and everything is ok.
I'm not sure this is the right way to describe what is happening. This "refine registration" algorithm adjusts a bit the registration based on the two sets of NAS/LPA/RPA points (one from the MRI, one from the digitized or MEG), at it does so using the head surfaces computed from the MRI + the digitized head points.
But you can't really say that before the sensors are in a "coordinate system based on the digitized anatomical landmarks" and after in "a MRI-based coordinate system". Both before and after, the alignment is the result of matching a set of points coming from the MRI and a set of points coming from the digitizer or the MEG.
Then warping displaces as well as deforms the MRI to match the head points, so that everything then really ends up in the digitized anatomical fiducials coordinate system.
This sounds ok.
So while both sets of fiducials won't be exactly the same, they should both define the same coordinate system axes.
But maybe there is something wrong with importing more CTF MEG recordings with a .pos file then warping?
Please let me know if you think there is anything that should be studied more in depth here.
Thanks Francois, I think we're in agreement conceptually, if not on how to describe it.
Before "refine", we have two independent sets of data (MRI and MEG/digitized) with coordinates each based on their own set of fiducial points. We're "matching" them simply by assuming that the digitized and MRI landmarks are the same. (This is different than if we did the same kind of point alignment with the 3 landmarks as with the head points for example.) So it is accurate at that point to say the sensor positions are in a coordinate system defined by the digitized landmarks. Their coordinate values don't depend at all on the MRI side.
Once we refine (ignoring how the alignment is done), they're converted to the "MRI-fiducial-based coordinate system". My point was that the MEG/digitized points coordinates are changed by the "refine" process, and not the MRI coordinates.
No, I think it's all good. Again I think if there's anything odd, it's in the real-time code, which I'll get back to shortly.
Hi François,
Sorry, but I'd like to bring this question of MRI vs digitized anatomical points up again... As I'm importing a few hundreds of participants in Brainstorm for the first time, I realized that it would be greatly advantageous to have the option to adjust the MRI anatomical fiducials to match the digitized ones after "refine using head points". Otherwise, one has to manually adjust the MRI fids for each subject, or accept that they won't be placed correctly, by the initial automatic MRI importing process. Is that correct?
I understand that in Brainstorm, head points are attached to single recordings, but that's ok. The matching I'm proposing would only have to be done once per subject after "refine using head points" on any recording. And even if it is repeated with another recording and the coordinates overwritten, it would either be the same coordinates (if same MEG session) or give similar reasonably good placement of anatomical points (if it's another MEG session).
I wanted to first check with you that this feature doesn't already exist, right? I think it shouldn't be too difficult to do since it's already possible to change the MRI fiducials manually. If you could let me know how you would go about doing this, to help me get started on the right track, it would be great. I would want to run this before exporting the co-registration for the OMEGA release, which I was hoping to finish this week.
Thanks!
I looked through the code and was surprised to find that adjusting the MRI fiducial points (nasion and pre-auricular points) breaks coregistration, if it was done with refine with head points. We might want to at least add a warning when modifying them, or if any transformations labeled 'refine registration: head points' exist, run it again.
That being the case, I'll implement my request as an option when refining with head points since it is a prerequisite and the transformation needs to be adjusted after changing the MRI anat points.
You should not modify the NAS/LPA/RPA points after any file beyond the MRI is imported.
The position of these points can be arbitrary (different conventions of anatomical identifications, HPI coils, adjustment due to hair, SEEG electrodes, etc...).
It you have enough head points to run an automatic registration, it does not matter where these points are at all.
Otherwise, one has to manually adjust the MRI fids for each subject, or accept that they won't be placed correctly, by the initial automatic MRI importing process.
Option B: accept that they are not placed "correctly", as they are no absolute "correct placement" anyway.
I wanted to first check with you that this feature doesn't already exist, right?
It doesn't and it won't. We don't want the anatomical landmarks to depend on any recordings. These are two separate processing streams.
I looked through the code and was surprised to find that adjusting the MRI fiducial points (nasion and pre-auricular points) breaks coregistration
Indeed.
We might want to at least add a warning when modifying them, or if any transformations labeled 'refine registration: head points' exist, run it again.
Done:
I spent the day yesterday implementing this so I'll post a pull request so you can have a look and we can discuss it. I still need to test and debug it today.
The reason I wanted it is for saving coregistration information in a large BIDS dataset (OMEGA). This is not required, but very useful since the MRIs will be defaced in the shared version, which would make it more difficult to use the head points for alignment. The way this coregistration is saved is by the coordinates of the anatomical fiducial points in both the MRI and functional file metadata. In my case, the latter come from digitization, so to provide the best alignment, I need these same coordinates on the MRI. As I'm typing this, I realize we don't need to apply them to the MRI in Brainstorm to save them in the _T1w.json BIDS file, but I still think it's a very nice feature to have. At the very least it looks better, and helps with quality checking.
In my case (import BIDS), both anatomy and functional files are imported together. And the functional files may be loaded first for different reasons, e.g. they are available first. Of course, coregistration should be done very early in the pipeline. This new option doesn't change that.
True, but I'm sure some people (like myself) won't be satisfied with anatomical points floating in mid-air or inside the head. I'm certain many hours have been spent by users manually placing these points. If they have a convention and they use it while digitizing, it just makes sense to me to use that available information and save the manual work. It has the added benefit of not having 2 distinct sets of anatomical points displayed on the "check registration" figure.
It's not a dependence, it's just a one-time adjustment of the MRI fids that's done automatically instead of manually.
One last note: The ChannelMat.SCS coordinates are are not currently adjusted when doing "refine with head points". I think it would be safer and more consistent to do it.
At the very least it looks better, and helps with quality checking.
What helps for quality checking is exactly to see that the two set of fiducial points (from the anatomy and from the sensors) do not match.
If you are planning on using the fiducials from the MRI, you should mark them manually on the MRI, or import positions that were previously saved. When we designed the data import pipeline for OMEGA with Guiomar, the data manager was supposed to mark manually these fiducials for each participant, and this info was saved in a file fiducials.m file (menu File > Batch MRI fiducials).
The manually marked fiducials should be saved in the BIDS metadata, and imported again when importing the BIDS dataset. This is indeed probably missing in the BIDS import/export at the moment.
If you are interested in developing code for improving the management of the fiducials for the OMEGA BIDS database, this missing feature would be a target of choice.
If you are planning on using only the automatic registration, then the information about the fiducials could be mostly ignored, as they are only a first step of initialization of the algorithm.
The keep on indicating, however, the starting point of the fit. This "automatic registration" is iterative and dependent on the initialization. Whether you start from two sets of point matching perfectly or from two divergent set has an impact on the quality of the registration.
In my case (import BIDS), both anatomy and functional files are imported together. And the functional files may be loaded first for different reasons, e.g. they are available first.
Then this is a special case, that needs to be addressed separately from the standard Brainstorm workflow.
True, but I'm sure some people (like myself) won't be satisfied with anatomical points floating in mid-air or inside the head.
Then mark them manually, as initially planned. This is what they've done in all the publicly available MEG datasets. Nothing will work better than this, and it would only take a minute per participant. Unbeatable in terms of result / time invested, for the limited number of participants in OMEGA.
I'm certain many hours have been spent by users manually placing these points.
In this case, they don't have this issue of MRI fiducials floating in the void.
This is only in the lazy case of using the default MRI fiducials from the poor linear MNI registration.
It has the added benefit of not having 2 distinct sets of anatomical points displayed on the "check registration" figure.
Having the two sets of points in the 3D view does have an important added value: it shows the mismatch between the two procedures. If there is a distance between the two versions of each fiducial point, it indicates that the two procedures of identification of the point are not leading to the same result.
It is an important information we definitely don't want to discard.
A good quality control for the MEG/MRI registration of the OMEGA participants would be to see the two sets of points matching very closely.
It's not a dependence, it's just a one-time adjustment of the MRI fids that's done automatically instead of manually.
If you don't want to mark manually the fiducials on the MRIs of the OMEGA participants before defacing AND don't want to see visually the mismatch of fiducials, then you need to develop an import pipeline that is specific for OMEGA. This might fit better as a dedicated import script than as part of the main Brainstorm distribution.
One last note: The ChannelMat.SCS coordinates are are not currently adjusted when doing "refine with head points". I think it would be safer and more consistent to do it.
The fiducials from the EEG/MEG/NIRS dataset are stored in the field HeadPoints, with type 'CARDINAL'.
The field ChannelMat.SCS is not part of the specification of the channel file:
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#On_the_hard_drive
It is some sort of hack that serves only one purpose, when importing for the time a data file that includes sensors positions + fiducials: transferring the positions of the fiducials from the low-level reading function (in_...) to the function that does an first level of auto-formatting of the channel file, which includes a first realignment of the sensors in a pseudo-SCS (based on the digitized fiducials instead of the anatomical ones). This happens here:
Instead of trying to maintain this redundant info, which would result in a lot of work for absolutely no outcome (as we really don't want to use this info anywhere but in channel_detect_type), if you want to clean-up the data structures, we'd rather delete the field ChannelMat.SCS after it is used in channel_detect_type.
Lets continue the discussion in the pull request.
However, regarding ChannelMat.SCS, if you plan on deleting it, please make the part that computes it into a function we can call. Since it is currently saved, I have used it.
You should not use this field, as you already noticed it is not updated with all the subsequent modifications of the channel file (including the automatic registration).
Please stick to the fields documented in the tutorials.