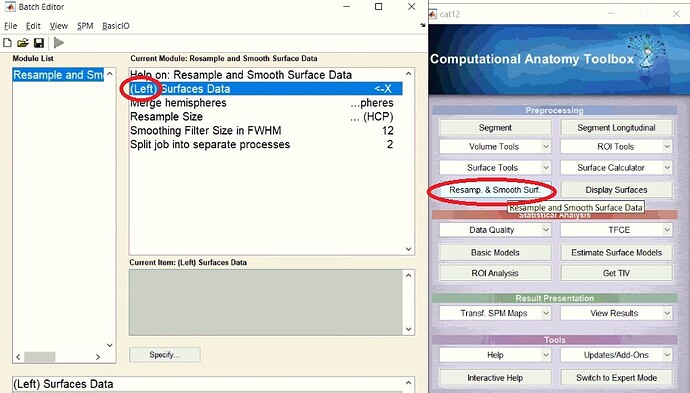
Hello everyone
I want to measure cortical thickness with CAT12 surface resample tool. But however I had both of ( lh.thick.thickness & rh.thickness) in "Surf" folder after doing "segment data" analysis, but when I want to enter my input ".thickness" datasets in surface tool,just
"left hemisphere.thickness" data can be loaded!!!!
Why "rh.thickness" data was not used in estimation of cortical thickness in CAT12 surface resample tool?
On the other hand I selected the merging hemisphere option in the section of "Merge Hemispheres".
How can I solve this problem?
Thanks
I'm not sure I understand what you are doing here.
From Brainstorm, select option "Import additional cortical maps" to compute the thickness maps:
https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12#Run_CAT_from_Brainstorm
When importing an existing CAT12 folder, select the file format "CAT12 + Thickness" to import the thickness maps:
https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12#Importing_the_results_in_Brainstorm
If your question is about the CAT12 interface, please post your question on the SPM mailing list.
If your question is about the Brainstorm interface, please be more specific of what you are doing: describe step by step what you are doing and include screen captures to illustrate each step.
Thanks
@CGaser FYI
Thanks for prompt response. Yes my question is about CAT12 interface and not about Brainstorm. Can you help me where I can write my question?
Thanks
@Mozhdeh123
In the newer version of CAT12 it's mentioned in the help text that the right hemisphere will be automatically processed. Thus, it's sufficient to select the left hemisphere data only.
Best,
Christian
Many thanks for your prompt response.
Hi CGaser,
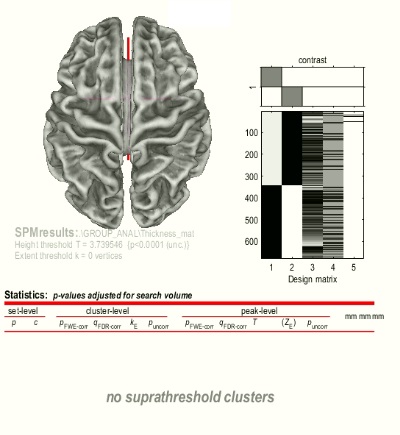
I implemented surface based morphometry on a datasets on which I
have previously performed VBM in CAT12/SPM12. The results for VBM are as expected but, for SBM, I am getting no suprathreshold clusters in both FWE & uncorrected p value!!! I was wondering because the datasets are similar in both VBM and SBM!!
Can you help me which step of simulation is wrong that I get this result?
@Mozhdeh123
Sensitivity for cortical thickness might be lower compared to VBM. However, I would first check the homogeneity of your data with the respective CAT12 tool to check for potential processing errors. Check the Quick Start Guide for the needed steps.
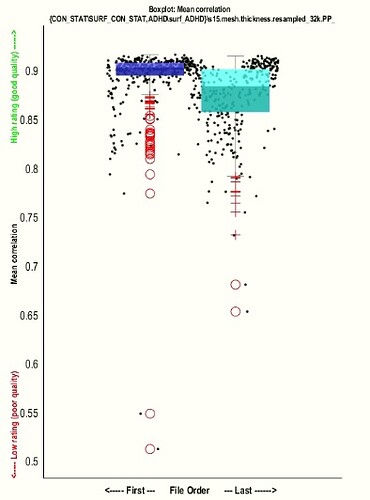
Thanks for your kind response. I checked smoothed thickness data quality with option of " check sample homogeneity of surface" in CAT12 such a way that I created two samples below Data and in sample 1 I uploaded the control groups smoothed thickness data and in sample 2 I uploaded patients group smoothed thickness datasets.after running this module results are shown like:
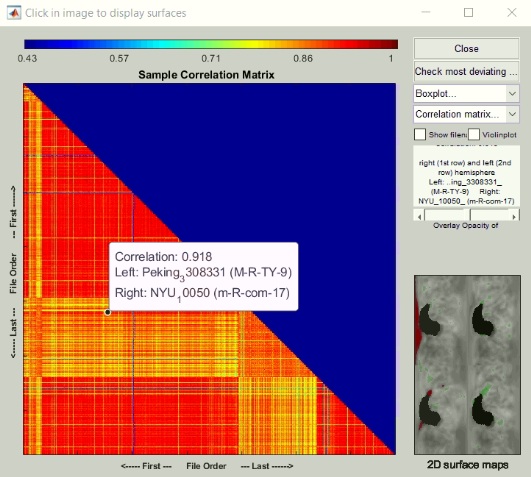
Given this result,are my datasets heterogeneous or no?
It is worth noting that just I changed the smooth kernel to 15mm and other settings in processes of extracting and smoothing &resampling of thickness datasets are the same as default.
- What could be the reason of this problem("no suprathreshold in cluster"??
- Which part of the default settings do you think should be changed?(I mean "resample size"?? or "FWMH kernel size"??? or other suggestions????
Thanks