Hi Santani,
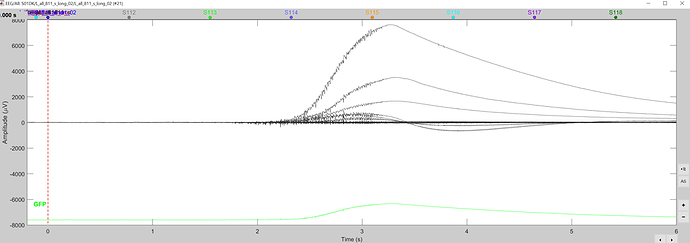
The problem in your data is that you have several bad trials, e.g. like this:
This causes havoc to the data normalization, which precedes the decoding estimation.
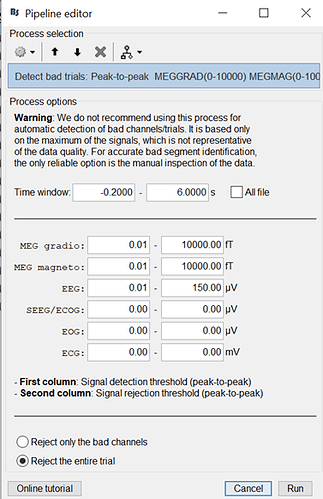
Use the following artifact detection process to exclude bad trials (notice 0.01 to 150 μV for your EEG data). This will discard about 10 our of ~80 trials per condition. Then decoding results will be similar whether you use whitening or not.
Notice, the functions you shared are old versions of my decoding scripts which I believe normalize data differently (e.g. use PCA). I highly advice you use the toolbox now distributed within brainstorm called 'scilearnlab' inside the 'external' folder. The function calls are easier now, e.g. like this:
d = sll_decodesvm(data,condid,'numpermutation',10,'verbose',2,'kfold',5, 'whiten', true);
Also notice, using the Brainstorm code, I got stepwise artifacts similar to yours when the option 'whiten' was true. This is because the whiten process fails with such huge artifacts. When disabling the whiten process (whiten = false), I did not get such artifacts. This is opposite to what you reported (in your case whiten works, non-whiten fails). But the discrepancy with your findings is because we are using different versions of the code with different normalization approaches.
Hope this helps!
Dimitrios