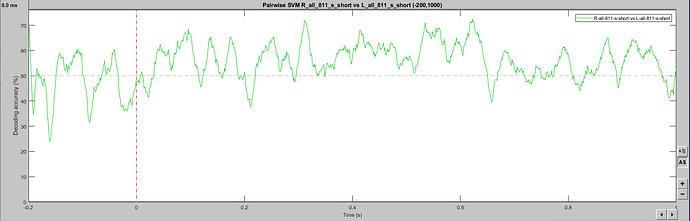
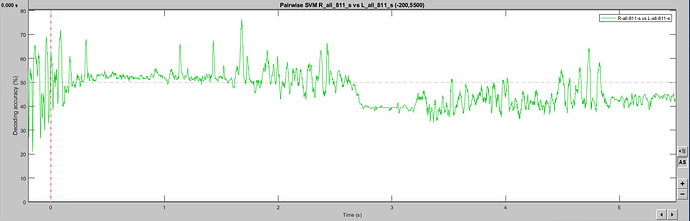
Hello, I may be missing something obvious, but SVM classification seems to change when a larger epoch is selected. I have two event classes being decoded in a [-200, 1000] epoch, which gives a noisy (but predicted) single-subject result. The exact same events imported with a [-200, 6000] epoch produce a drastically different outcome, including within the [-200, 1000] window. As I understand the temporal decoding process, that should not be the case. Is this a bug? Feature? User error? Thanks for any insight! This is 64ch actiCHamp EEG data, with about 80 trials per condition. Baseline-normalized and 30Hz filtered.
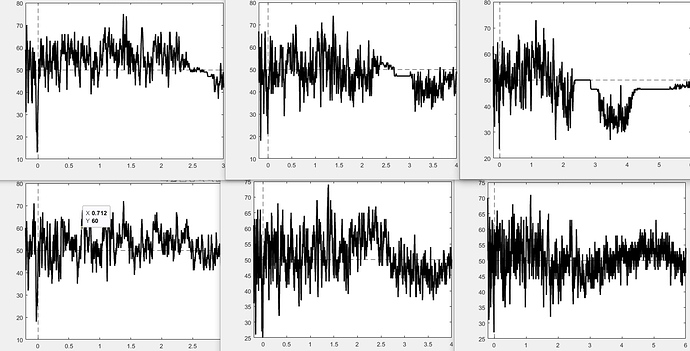
As an update, I exported the trials and ran the decoding externally. The top row of the new attachment is the 1d svm decoding at 3, 4, and 6 s. The bottom row is the same but with whitening turned off. So it looks like something about the noise normalization breaks after an epoch length of ~2-2.5 seconds: Is there a workaround aside from disabling that function?
This is not a deterministic process, there are some random grouping of trials: https://neuroimage.usc.edu/brainstorm/Tutorials/Decoding#Decoding_with_cross-validation
Re-run the same process multiple times on exactly the same data, you'll get different results.
Examples with the tutorial dataset:
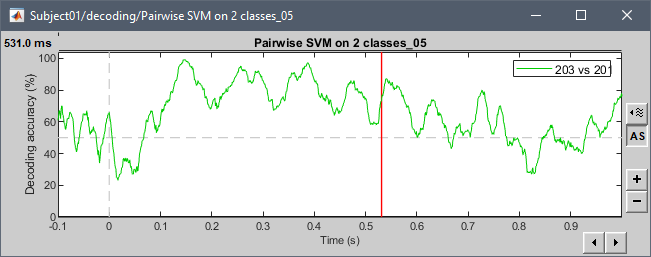
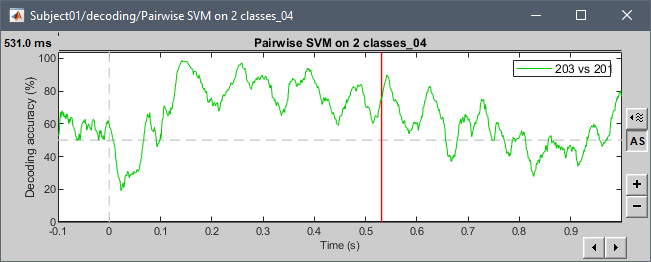
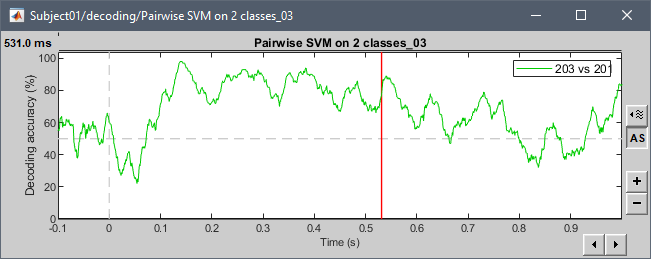
@pantazis Any additional comments?
Thank you for the response! I understand the normal variation from the permutation/subaveraing process, but this is way out of the range of that expected variation. The weird feature ≥2.5sec is very reliably present with vs. without whitening.
Hi Santani,
This appears to be a very peculiar problem. To investigate, can you please send me an email (pantazis@mit.edu) with a link to your data to replicate the problem? It can be either a brainstorm database, or the exported data directly used by the decoding functions.
Best,
Dimitrios
Thanks Dimitrios! I've sent you a separate link.
Hi Santani,
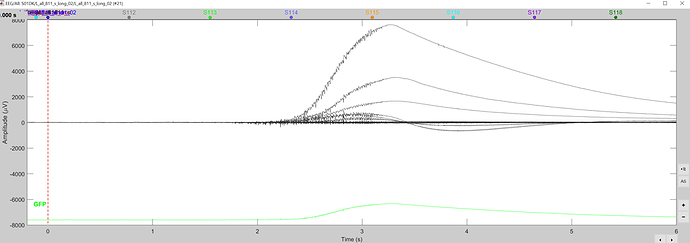
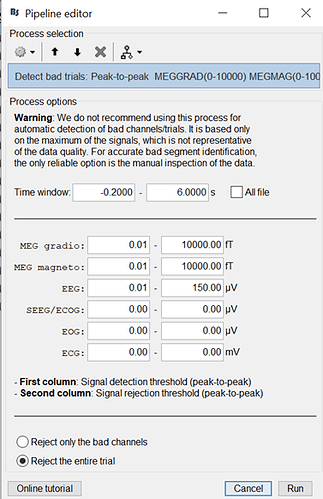
The problem in your data is that you have several bad trials, e.g. like this:
This causes havoc to the data normalization, which precedes the decoding estimation.
Use the following artifact detection process to exclude bad trials (notice 0.01 to 150 μV for your EEG data). This will discard about 10 our of ~80 trials per condition. Then decoding results will be similar whether you use whitening or not.
Notice, the functions you shared are old versions of my decoding scripts which I believe normalize data differently (e.g. use PCA). I highly advice you use the toolbox now distributed within brainstorm called 'scilearnlab' inside the 'external' folder. The function calls are easier now, e.g. like this:
d = sll_decodesvm(data,condid,'numpermutation',10,'verbose',2,'kfold',5, 'whiten', true);
Also notice, using the Brainstorm code, I got stepwise artifacts similar to yours when the option 'whiten' was true. This is because the whiten process fails with such huge artifacts. When disabling the whiten process (whiten = false), I did not get such artifacts. This is opposite to what you reported (in your case whiten works, non-whiten fails). But the discrepancy with your findings is because we are using different versions of the code with different normalization approaches.
Hope this helps!
Dimitrios