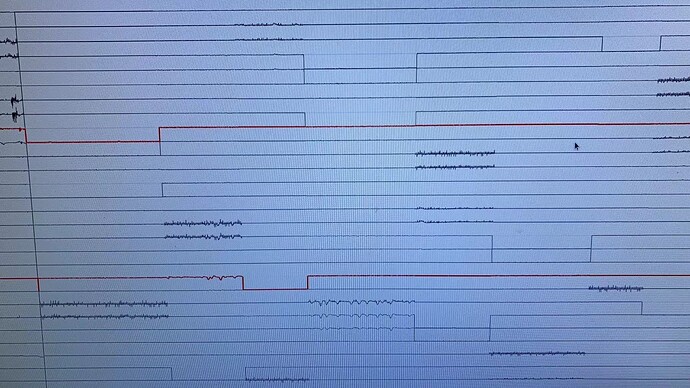
I use brainstorm to read data from Nihon Kohden EEG-1200A, system. The data show normal in the Nihon Kohden review software, but the data does not appear to be normal after import from brainstorm. The data were successfully imported. I attached one picture I took from brainstorm eeg viewers. Many channels do not have values. Do anyone know potiential bugs in the io script?
Unfortunately, we don't have access to the specification of this file format, and Nihon Kohden doesn't provide any library to read their files...
If you shared a short example file with me, I could give a try in understanding what the problem is, but I don't have much hope.
If you need to access this file in Brainstorm urgently, you can export it to EDF+ file format from the Nihon Kohden software.
I shared two files in the following google drive link. One is the raw eeg file from NKT system, and the other is the ASCII file for the same data. I need to import large amount of data, and hence export each data one-by-one is time consuming. Please help me to see how to import directly from brainstorm.
I tried to debug the code from Brainstorm. It seems that one of the block address was read wrongly. Therefore, the total length of the data are wrong. You could use the default 10-20 channel names.
https://drive.google.com/drive/folders/1Kd0je5yrnU0LWbSxXTT-NvwWLt42qAA1?usp=sharing
I can't download the file, the access is restricted.
Now should be fine. Sorry for the inconvenience.
Thank you for the example file.
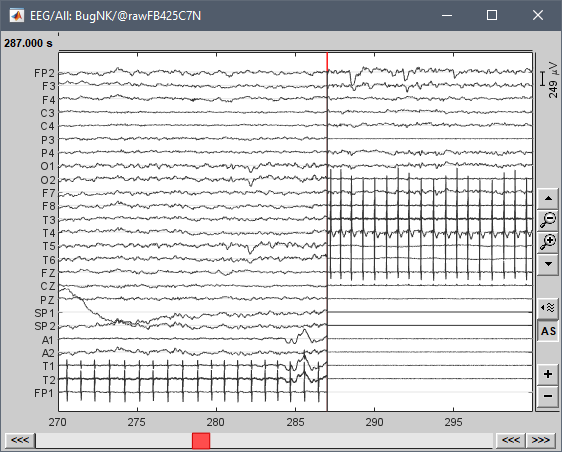
The recordings look normal until 287s, and then it looks like there is some shift in the data matrix.
From what we read in the header, it looks like there is only one control block and one data block:
>> hdr.ctl
struct with fields:
address: 1024
data_cnt: 69
data: [1×1 struct]
extblock2_address: 9263
>> hdr.ctl(1).data(i)
struct with fields:
address: 6142
timestamp: 16885
sample_rate: 500
num_records: []
num_samples: []
extblock3_address: 15403
num_channels: 44
channel_list: [1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 43 44 45 46 47 48 49 50 77 78]
rec_address: 15905
There is one unclear definition, which is the duration of this data block. It is not defined in the header, therefore our reading function tries to guess it from the size of the file:
This gives a duration of 393011 samples / 786.02 seconds.
However, this is probably not reflecting the actual size of the first block. One hypothesis could be the following: the first data block is only 287s long and it is read correctly. But the file contains more recordings for which Brainstorm doesn't have any entry point (the header is most likely read incompletely). When detecting the first block duration, it uses the file size, which includes a lot more recordings than only the first block. But after 287s, Brainstorm reads data from a wrong starting point and therefore the EEG data is all messed up.
Unfortunately, I don't know how to go further in debugging the reader with this file...
If you have access side-by-side to a viewer that can read the signals beyond 287s, you could: first make sure that the beginning of the file is read correctly, then try to understand where to find the address and description of the other data blocks in the file...
Otherwise, if you find an open-source software that can read this file correctly past 287s, we could explore the code and fix the Brainstorm reader.
Don't count on the NK customer service: they are more likely to send you a lawyer for breaking the user agreements, than an engineer to help you work with the data...
Last solution: Export the file to EDF+ from the NK software (or other programs that have a non-disclosure agreement with NK, like BESA or Curry), and then work in Brainstorm with the EDF files.
Thanks for your response. Yes, I found the same issue as you did and cannot figure out how to deal with it. I finally used NK software to export the data, but it is quit time consuming. Thank you all the same for trying it.
Yours,
Hi Francois,
From your experience, how could we select the rank of the lead field matrix when we do the source localization. I found that the rank can affect results significantly.
For each new topic, please create a new thread on the forum.
You can't control directly the rank of the lead field matrix from the Brainstorm interface.
If you want to see how this is estimated in the code, see: