Yes, of course!
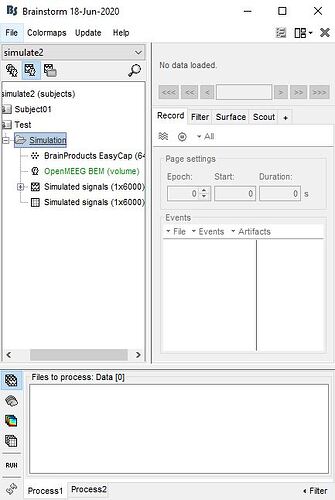
I'm going to simulate EEG recording by myself in Matlab. For this objective, I've used defult anatomy in BS software and defined:
a point scout (seed:12497) as a source
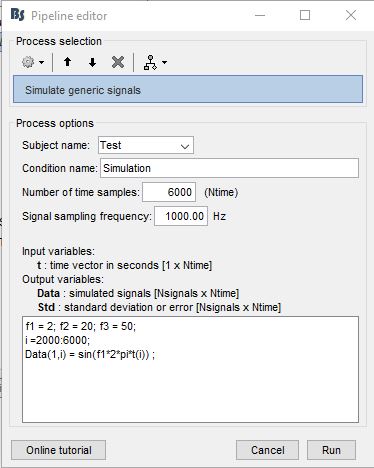
a simulated generic signal as:
f1 = 2; f2 = 20; f3 = 50;
i =2000:6000;
Data(1,i) = sin(f12pi*t(i)) ;
and assigned this signal to the scout. Also, I've used BS's OpenMEEG BEM (volume) to calculate the head model. In this step, "I asked BS to simulate recorded signals in the default 64 channel cap".
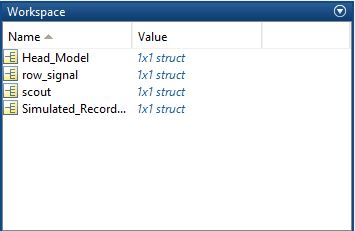
I want to do this step (simulate recorded signals in the default 64 channel cap) by myself in Matlab. To do this, I've exported the following cases to Matlab:
-
scout seed number as "scout"
-
simulated signal (Data(1,i) = sin(f12pi*t(I)) ) as "row_signal"
-
OpenMEEG BEM (volume) as "Head_Model"
-
simulated signal as "Simulated_Recording" for result comparing.
for simulating recorded signal, I need to know how I can multiple Head_Model with row_signal. @SBeumer gave me very good tips but I've not succeeded to have the same results those are shown by Simulated_Recording yet.
In other words, my problem is how I can convert "Head_Model.Gain" with 64 * (3 x vertexes) in 3 dimensions (x,y,z) to a Head_Model matrix with 64*vertexes.
I hope to describe my problem better
Regards
Morteza