Hello,
I try to apply time-frequency analyzes on SEEG data. But I only get results on electrodes (N1) whereas I would like to have an analysis on bipolar electrodes (N1-N2)... Probably not a big question but I can't find the solution... Thank you
Hi Florian,
This can be done by using montages: https://neuroimage.usc.edu/brainstorm/Tutorials/MontageEditor
More specifically, you will need to create a custom montage with the desired biopolar channel:
N1-N2 : N1, -N2
After you need to apply the montage to the recordings with the process Standardize > Apply montage. This will create a file with the N1-N2 channel, then you can proceed with the TF analysis as usual
Best,
Raymundo
That's work, thank you. I had missed the last step.
F
Hi Florian,
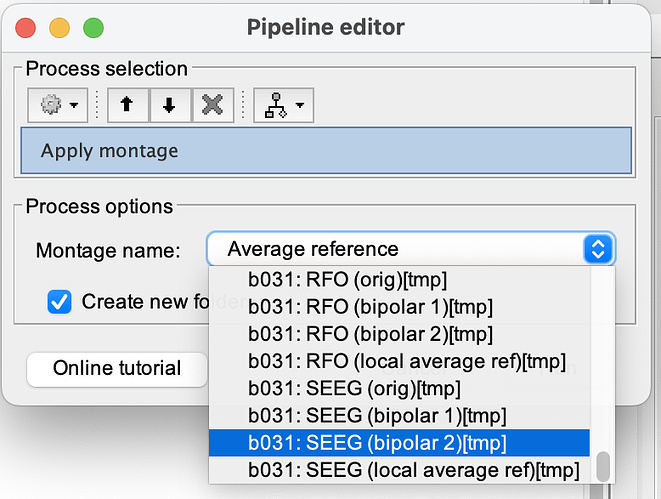
I have done this without making a custom montage. If you have correctly labeled all of the SEEG contacts as SEEG in the channel file, brainstorm creates temporary bipolar SEEG montages for you. It should look something like the attached picture (: SEEG (bipolar2) [tmp]). These montages are subject-specific, because each subject may have a different electrode array. Bipolar 2 is the more complete montage in which adjacent channels share an electrode in common (ie. A1-A2; A2-A3; A3-4, etc). Bipolar 1 is the more sparse montage where there are fewer channels and adjacent channels do not share any common electrodes (A1-A2; A3-A4, etc). This is extremely helpful if you have a lot of subjects.
Tip: You need to import to the database before you can use the "Apply montage" process. Although you can view raw files in the bipolar montages, the "Apply montage" process is grayed out for raw files.
Best,
Dan
Thank you for the details, Dan!