I am a beginner, Please help me with this trouble, thx!
I imported a cnt file but brainstorm couldn’t recognize it. Then I reimported the location file(Neuroscan_Quik_cap64), it still didn’t work after the calibration
. It shows as an electrode which only represents half of it, and cannot read the data properly. So could you guys tell me how can I solve this problem? Thx a lot.
Matlab version: 2013a
Brainstorm version: up to date(Brainstorm 17-Mar-2017).
Hello,
Have you followed the introduction tutorials (in the section “Get started”) and then the EEG/Epilepsy tutorial on the website?
http://neuroimage.usc.edu/brainstorm/Tutorials
Is your problem that half the signals are shown as flat (= zero) in the figure with all the signals?
Can you describe exactly what is different between what you expect and what you actually see in Brainstorm?
If you cannot find yourself the solution with the tutorials, can you please share this file so we can look at it?
(upload this file somewhere, eg. dropbox, then send me the link in a separate email - click on my username on this forum)
Francois
Hi Francois, thx for your notice. What I actually see in Brainstorm is that there are a half of channels that cannot be read, and it shows a straight line. What I expected was that brainstorm could read the information of all the channels rather than that of only half of them. I cannot perform the following operations, and I have no idea whether it is the problem of importing the location file or not.
And there isn’t any detailed guidance of how to change channel file (EEG) in the tutorial.
I’ve sent you an email in which includes the link of .cnt file that I have trouble with. Thx!
Hello,
Thank you for the file.
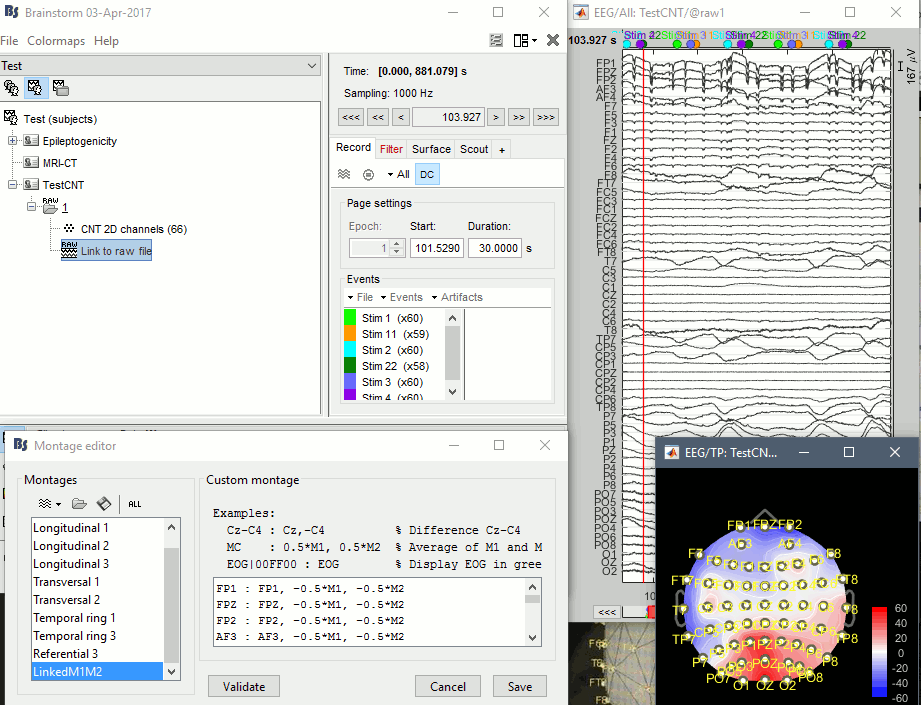
I'm not sure I understand how to got to the screen captures you sent me. There is no problem with displaying or processing this file (see attached screen capture)
What I did:
- Create a subject (one channel file per folder), right-click > Review raw file
- Right-click on the channel file > Add EEG positions> ICBM152 > ASA 392 (your file should still be named "CNT 2D channels", unlike the screen capture your posted)
- Right-click on the channel file > Edit channel file > Change the type of M1 and M2 to "REF") => It produces nice 2D topographies
- Created a new montage with linked mastoid references (M1+M2) => Illustrated in the screen capture, but useless because I guess the recordings are already correctly referenced to (M1+M2)/2
Have you computed ICA projectors that could have removed a lot of the signal (check in tab Record, menu Artifacts > Select active projectors)
If you still get these weird plots with every other channel that is flat, I'd recommend you delete your subject and start over from the beginning.
And make sure you work with a version of the Brainstorm that is up to date (menu Help > Update Brainstorm).
Cheers,
Francois
Hi Francois,
I reseted brainstorm and reperformed the import process from the beginning, but the same problem that half of the signals couldn’t be recognized happened again. Comparing my steps with yours, I found that the main problem may be the location file after the step that the cnt file was imported. According to your instruction, the CNT 2D channels(66) file may appear, but what I’ve got was only ANT standard position by default which would not change after I imported the location file of NeuroScan64 which is the brand of our electrode cap.
And then I updated the brainstorm to the up-to-date version 04-Apr-2017, there still existed the same problem, and couldn’t find the option of “ASA 392”, either, and I’ve uploaded two screenshots about it.
How can I solve this problem? Thx a lot!
If you get a file named like this, I think it is because you don’t follow this instructions correctly.
What you do is: right-click on the folder > Use default EEG cap.
What you are supposed to do: right-click on the the channel file (named initially “CNT 2D channel”, right after you link the recordings to the database) > Add EEG positions.
It was not “ASA 392” but “ANT 343”, sorry.
Hi Francois,
Thx for your instructions, but maybe you got me wrong. I am certain that I have followed yours correctly.
Just after I selected the option “Review raw file” from step 1:“1) Create a subject (one channel file per folder), right-click > Review raw file”, a dialog box popped up asking me to select a cnt file, and after I selected the cnt file which I have sent you via google drive and clicked the “Choose” button, the system automatically showed the channel file named “ANT standard position (66)” below the Subject01 rather than “CNT 2D channel” without any notice asking me to select between them, and I didn’t do the step as you said of “Use default EEG cap”.
So could you tell me how to change my imported cnt file into “CNT 2D channel” file? Thx a lot!
Oh I just realized this is an EEProbe .cnt file, not a Neuroscan .cnt file…
I haven’t tested much the EEProbe reader, maybe there are issues with it.
I rely on compiled code from EEGLAB to read these files, there is not much I can do about it.
But I just noticed that the version of this library that is used in Brainstorm is not be the most recent one.
Could you try replacing the files in brainstorm3/external/eeprobe with the ones downloaded from the EEGLAB website?
http://sccn.ucsd.edu/eeglab/plugins/ANTeepimport1.13.zip
If this solves your issue, I will upgrade to this last version.
Thanks
Francois
Hi again,
I upgraded the version of the EEProbe library in Brainstorm, because it looked like it could read the test files I have faster.
To test this new version: simple update Brainstorm (menu Help > Update Brainstorm), delete the links to the .cnt files from your Brainstorm database, and create them again.
If it doesn’t work and you still get the same results, please contact the EEGLAB or ANT support.
I don’t have the corresponding source code and cannot help you further.
Cheers,
Francois
Hi Francois,
I’ve replaced the files with the ones you gave me, but there exists the same issue.
And after I upgraded Brainstorm to the latest version 17-Apr-2017 and tried again, nothing has changed.
Maybe it’s the type of the cnt file of mine that doesn’t match the CNT 2D channel type?
Or can we solve the problem that half of the signals are shown as flat (= zero) in the figure without the step of matching the type of CNT 2D channel?
Can you send me a copy of your Brainstorm program in your PC which can correctly open the cnt file I have given you without the problem? Thx!
Hi Francois,
The latest version works very well, and I uploaded the picture to show the result.
Thx for your great help, I may ask you for help if I encounter another problem.
Thanks for this thread. I’m also a newbie and using Neuroscan Curry 7 (38-channels) but can’t seem to find the 32-cap in Brainstorm. Failed uploading the file (.dat). Help please.
Cheers
What is the name of you EEG cap?
If it is a standard 10-20 placement, you can use the default positions available in the menu: Add EEG positions > ICBM152 > Generic