Hello,
Thank you for the file.
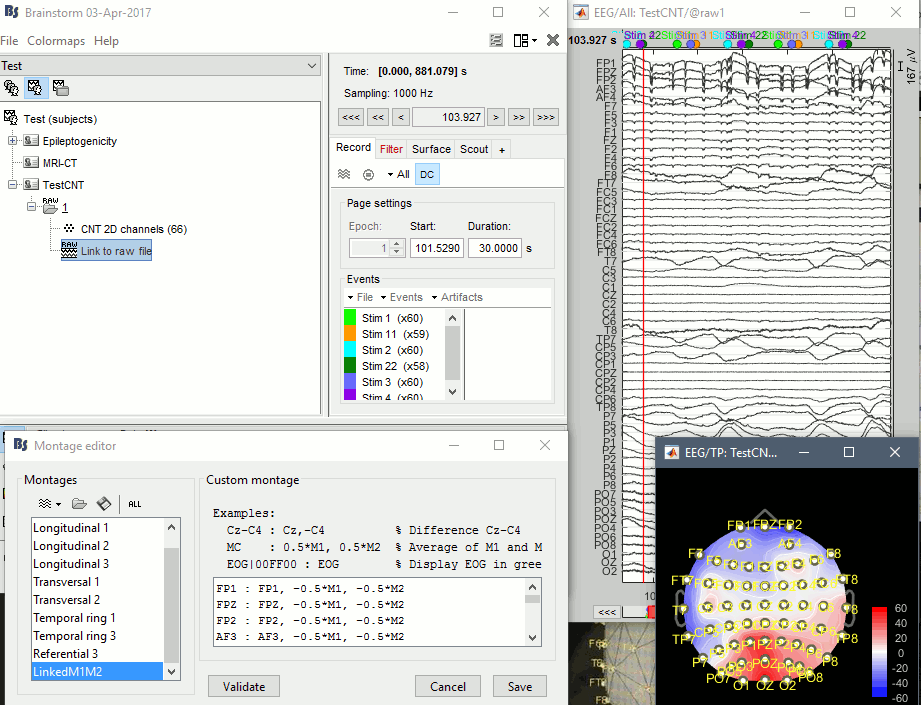
I'm not sure I understand how to got to the screen captures you sent me. There is no problem with displaying or processing this file (see attached screen capture)
What I did:
- Create a subject (one channel file per folder), right-click > Review raw file
- Right-click on the channel file > Add EEG positions> ICBM152 > ASA 392 (your file should still be named "CNT 2D channels", unlike the screen capture your posted)
- Right-click on the channel file > Edit channel file > Change the type of M1 and M2 to "REF") => It produces nice 2D topographies
- Created a new montage with linked mastoid references (M1+M2) => Illustrated in the screen capture, but useless because I guess the recordings are already correctly referenced to (M1+M2)/2
Have you computed ICA projectors that could have removed a lot of the signal (check in tab Record, menu Artifacts > Select active projectors)
If you still get these weird plots with every other channel that is flat, I'd recommend you delete your subject and start over from the beginning.
And make sure you work with a version of the Brainstorm that is up to date (menu Help > Update Brainstorm).
Cheers,
Francois