Hello! Has anyone been able/can you upload raw imaging data into brainstorm and convert to freesurfer file within the program? our lab just has dicom images (no nifti files yet), and we were wondering if that was possible to do that and move forward.
to note--our lab runs on Windows processing.
Hi @kjgrf
Yes, you can import Dicom files to brainstorm, and you can then export them to Nifti format.
you can also perform segmentation of the MRI using one of the plugins [Brainsuite, SPM, SimNibs ...]
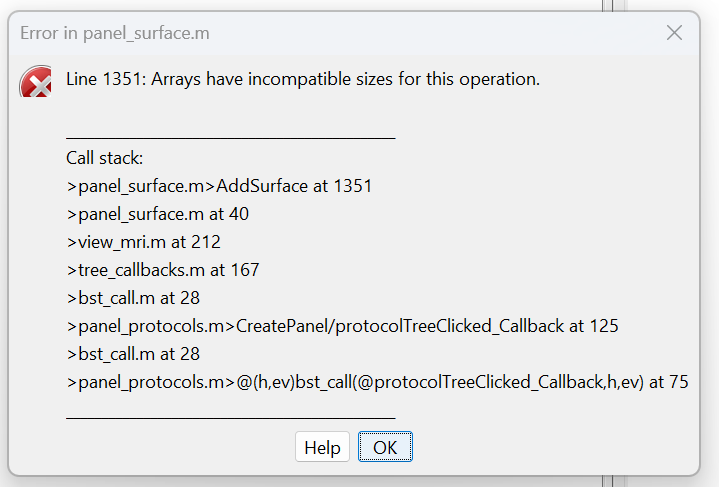
i was able to import a dicom file using the cat12 toolbox in spm, but it says there are mutliple issues with the arrays and I cannot view the MRI, any help?
I don't know what is this issue from this panel,
were you able to load the MRI to Brainstorm?
is the MRI icon visible in the anatomy panel?
you can also try to use the MRIcron,
https://www.nitrc.org/projects/mricron
it allows you to load and visualize dicom and then convert to nifti
Unfortunately, all of our raw data is unrecognizable in the dcm format, so having to convert all to dicom before I can convert to nifti. Since that may take a while, do you know how to run freesurfer through matlab on windows, is that even possible?
You can not run Freesurfer on Windows; Freesurfer is developed to work on Linux.
One possible solution is to use Windows Subsystem for Linux (WSL) or install a virtual machine on Windows and then install and run Free Surfer.
The other alternative is to use different software that runs on Windows, such as Brainsuite or SPM.
I am now stuck in uploading EEG data, i have it saved it a .mat file type, but I cannot load it onto brainstorm because it says there is no data to be read.