Dear Brainstorm community,
So far, I've been manually using the GUI to import individual EEGLAB files which are already preprocessed and epoched. The data is an ERP study, with 4 conditions of interest, with the event values of interest under EEG.event.type being 'Go_Happy', 'Go_Angry', 'NoGo_Happy', and 'NoGo_Angry'.
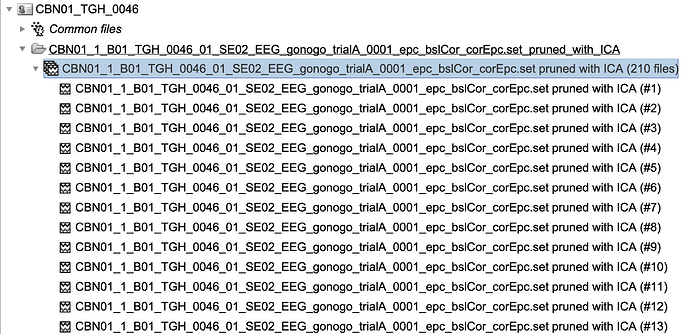
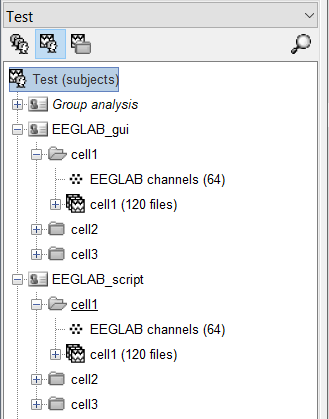
When using the GUI to import, I can select these 4 conditions, and Brainstorm imports the epochs into their own unique event-named folders like this:

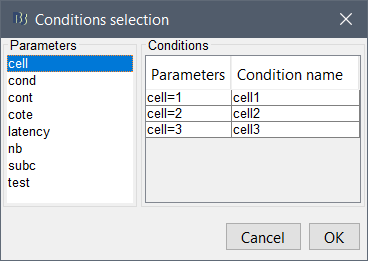
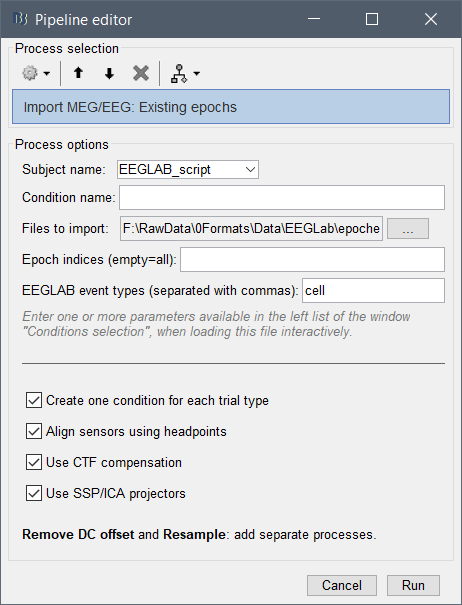
I would like to however script this process using 'process_import_data_epoch'. I have tried many combinations of filling in the 'eventtypes' field, but I keep ending up with a generic folder with all the epochs under it like this:
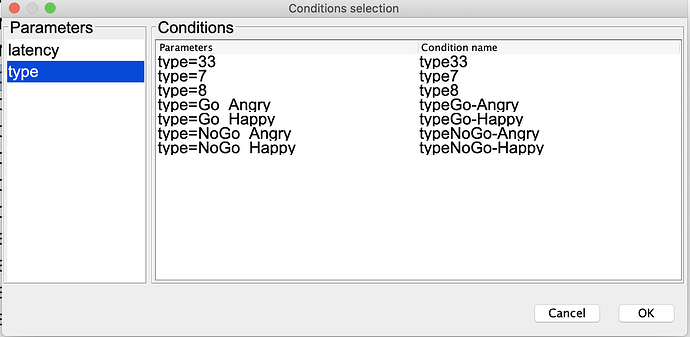
This is my Conditions selection screen when using the GUI:
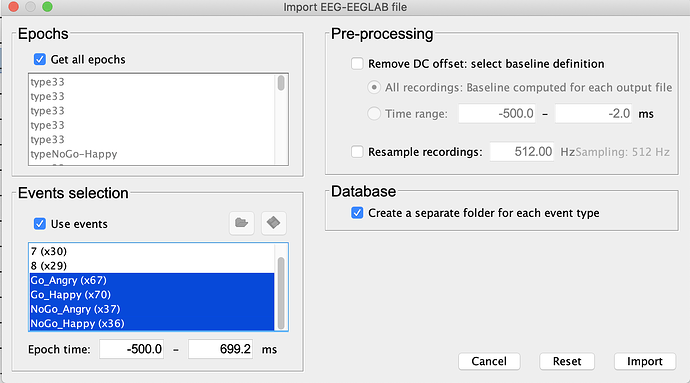
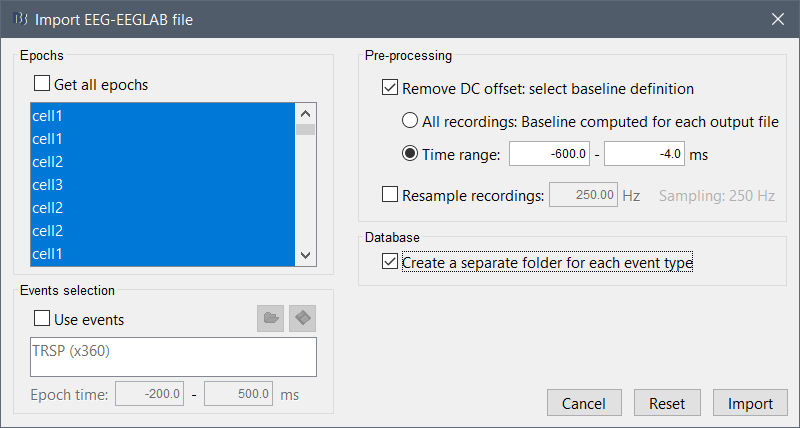
and the Import EEG-EEGLAB file screen:
Any help as to what I can do to get the epochs into their individual condition folders while using the script would be greatly appreciated.
Thank you,
Paul