Hi all,
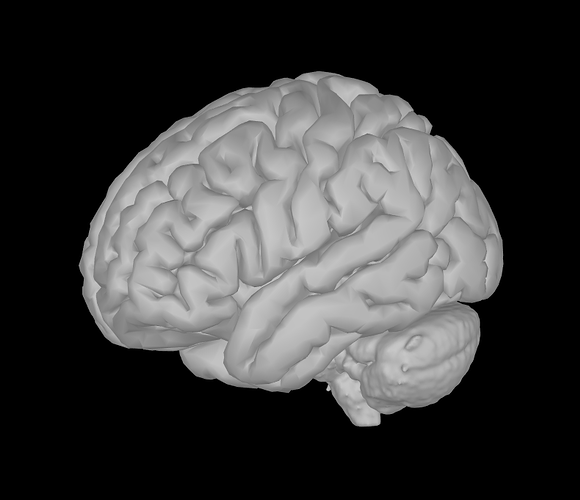
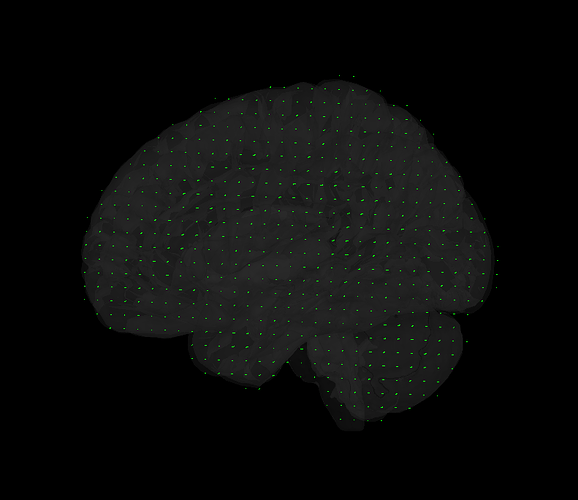
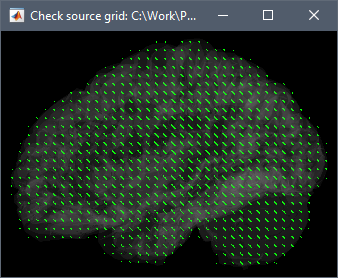
I'm interested in volume source solutions that include the brainstem. Only with MEG now. In the anatomy folder, I mixed a cortical surface with the aseg atlas to create a surface of the whole brain, brainstem included (picture). Having that set as "default cortex", I proceeded by generating a source grid/isotropic from the common files functional tab and, upon inspecting the grid, I believe I can see vertices in areas including the cerebelum and brainstem (picture 2). However, when generating the head model with the "use template grid for group analysis" option and the inverse solutions, the sources I get completely ignore areas outside the cortex. Brainstem and cerebelum areas are not even displaying values, so I am wondering why this happens.
Thank you a lot,
Fran
Hi @FranLop89
I think that the option "use template grid for group analysis" will interpolate the results that you have computed on your model to the "template cortex" which is not the one that you are using.
The template used in Brainstorm is the ICBM, this tuto may explains your issue:
https://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Project_sources_on_template
A+
Thank you @tmedani
I am not sure I understand your answer, I am not projecting the sources to a common space yet, I am having trouble seeing subcortical sources at the individual subject level, despite using a grid in which there are vertices in that area. This is the tutorial I was following: https://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects#Volume_source_models
Edit: upon close inspection, I can tell I have a value for each vertex in the volume grid if I open ImagingKernel variable in that subject. However, when I display the sources over the MRI only the cortical ones are displayed (have colors). How can I make it so that I see, for instance, the brainstem there as well?
Thanks again,
Fran
Are you sure you've selected the correct head model before running the source estimation?
Have you lowered the amplitude threshold (Surface tab) to make sure the deeper values are not simply lower?
Please post screen captures illustrating exactly your concerns.
And please include screen captures of the database explorer showing all the files involved (anatomy, headmodel, source results)
Thank you Francois,
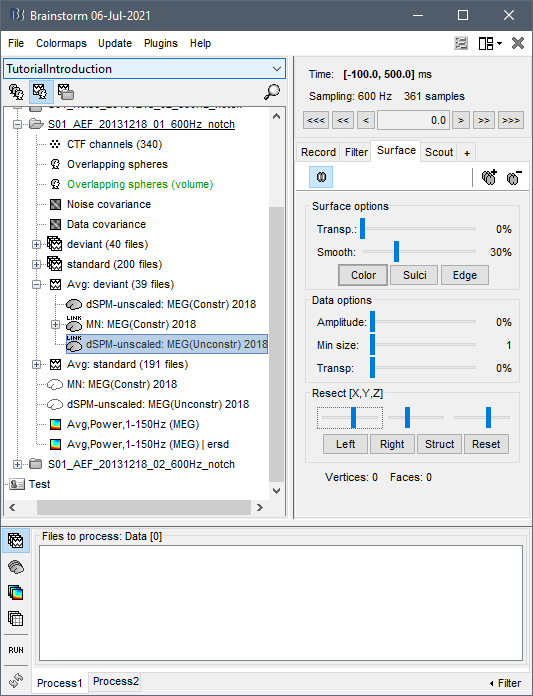
Apologies if I did not explain this clearly. Yes, I am sure the head model I want to use is in green before I run the source estimation. Here are the screen captures. The picture of the grid now is from the head model of that subject specifically (right click on head model/check source grid) and, as you can see, there are vertices in the area of the cerebelum for instance. However, when I right click into the source file (database picture), the option to display sources is "cortical activations/display on MRI" and, as the name states, it only displays the sources from the cortex (blurry picture). How can I make it so that I see source activations of the subcortical structures as well?
Thanks a lot,
Fran
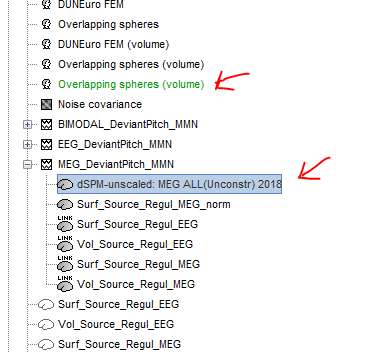

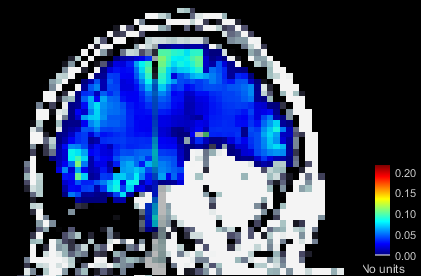
This should be working correctly, I guess there is something wrong in your setup.
What you see in your source map is coherent with what you see in the grid: the default cortex (selected in the green in the database explorer) does not include the cerebellum.
- Make sure your edit cortex (cortex+subcortical structures) is selected and displayed in green in the anatomy of your subject,
- Restart Brainstorm (to make sure no residual interpolation matrix remains in memory)
- Compute again the forward and inverse models.
If I reproduce what you describe on the introduction tutorials dataset, I get this:
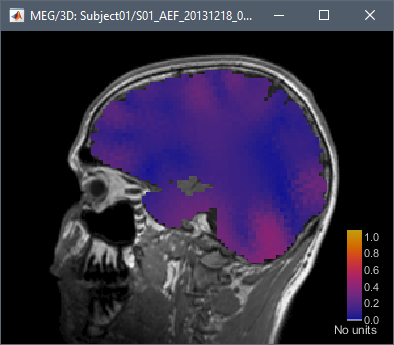
1 Like