Dear All,
I am doing resting-state MEG sensor-level analysis.
I have the subjects' Neuromag raw.fif files (tSSS-processed with the Neuromag MaxFilter right after MEG acquisition) with 3 fiducials, 4 HPI coils, and about 100+ Polhemus digitized head points. I do not have the subjects' T1 anatomy.
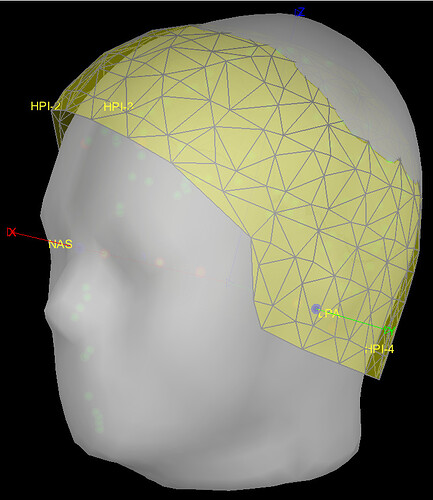
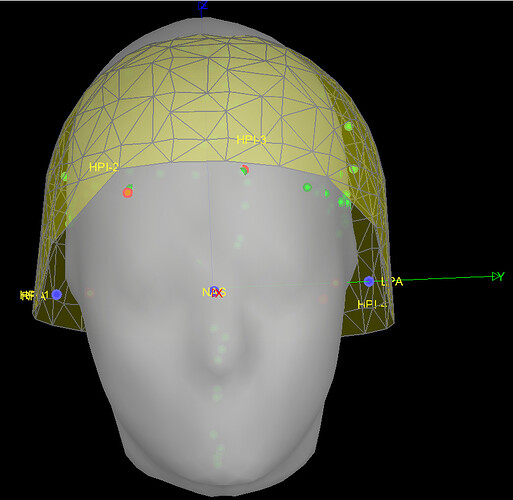
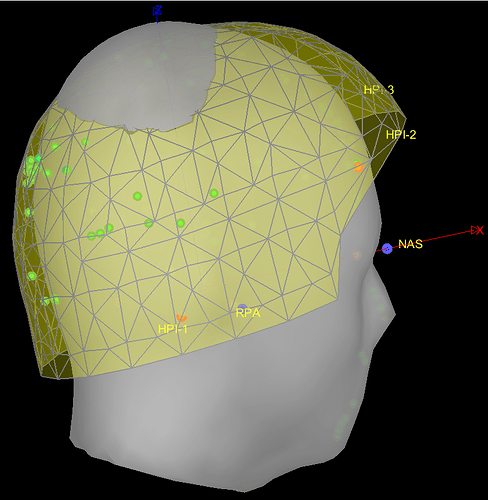
I'm currently encountering registration problems when trying to match the MEG helmet (yellow grids), the MRI template surface (grey), the subject's digitized head points (green dots), the subject's 3 fiducials (red dots), and the MRI template's fiducials (blue dots).
Basically, I follow the tutorial:
(A)
New Subject > Yes, use default anatomy > No, use one channel file per acquisition run (MEG/EEG) > Review raw file > Ignore (because I'm using resting-MEG) > Neuromag channel right-click > MRI registration > Check
In most of the individuals, the template head (grey surface) often intersects with the MEG helmet (yellow grids) as in the example below.
BEFORE (example):
I also checked the points in the "Anatomy" tab > Default anatomy > ICBM152 and confirmed that subjects' 3 fiducials are the same as the red dots in Brainstorm SCS.
Currently, I chose NOT to edit the points manually (in "Functional" tab > channel right-click > MRI registration > Edit) because I found it very hard to control the translation or rotation to get the subject's points/fiducials matched to the template's surface/fiducials.
===
(B) Subsequently, I proceed to the warping.
(B-1)
Channel file right-click > Digitized head points > Warp > Deform anatomy to fit these points > Warp > Remove outliers (2%)
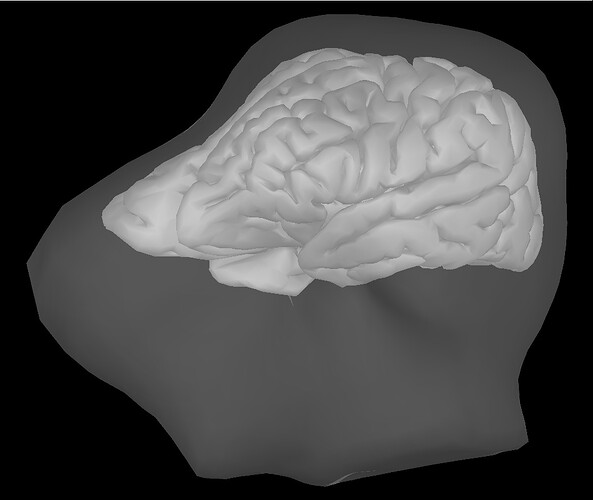
AFTER "Warping" (example): ![]()
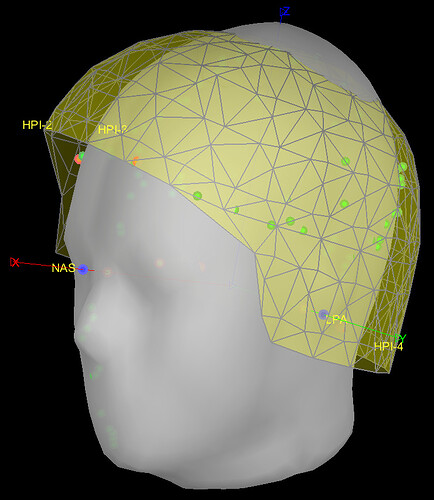
(B-2) But if I choose "Scale" instead of "Warp", the results seem a lot normal.
Channel file right-click > Digitized head points > Warp > Deform anatomy to fit these points > Scale > Remove outliers (2%)
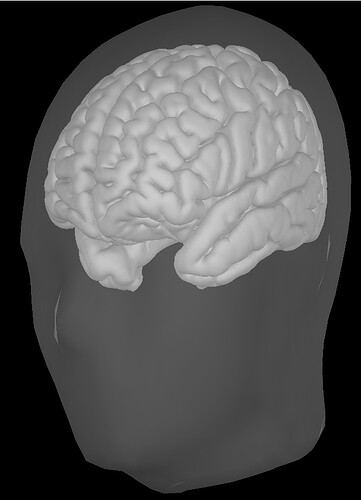
AFTER "Scaling" (example): ![]()
Although the template head surface still intersects with the MEG helmet, they looked a lot better and normal.
Therefore, I think I will use the B-2 method (only Scaling and not Warping).
But I wish to check with you guys if this is the correct way. Please advice. Thank you very much!