4th of September Brainstorm introduced support for fooof toolbox.
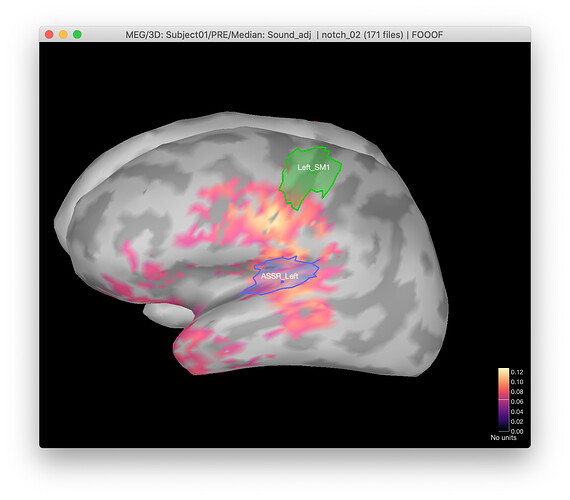
In my current pipeline, I was extracting values from fooof model from reconstructed sources. When running it first time by hand I was able to obtain results that make sense when visualizing the location of the peaks from fooof output (below is response to the stimulation).
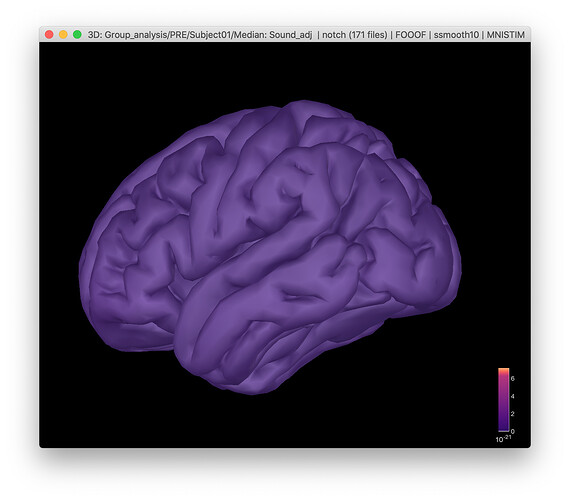
Now when I am trying to reproduce the results, I am getting strange-looking brains.
This picture was made with a setting for 0% amplitude, and the activation disappear when the setting is higher. When I try to plot single spectra or model values they are the same across the cortex. The saved fooof-output file seems to have different values for every source, so it seems ok.
Other times fooof peaks showed up all along with the frequency of the dataset (the fooof is fitted between 2-48, the peaks appeared up to 300hz, it looked like spectrum values were displayed instead of peaks.
Maybe the problem is how I extract PSD for fooof?
In my analysis, I should get the average/median of all spectra across trials, on for every reconstructed source and then I feed the spectra into fooof model.
MY code
% Process: Compute sources [2018]
sFiles = bst_process('CallProcess', 'process_inverse_2018', sFiles, [], ...
'output', 1, ... % Kernel only: shared
'inverse', struct(...
'Comment', 'MN: MEG', ...
'InverseMethod', 'minnorm', ...
'InverseMeasure', 'amplitude', ...
'SourceOrient', {{'fixed'}}, ...
'Loose', 0.2, ...
'UseDepth', 1, ...
'WeightExp', 0.5, ...
'WeightLimit', 10, ...
'NoiseMethod', 'median', ...
'NoiseReg', 0.1, ...
'SnrMethod', 'fixed', ...
'SnrRms', 1e-06, ...
'SnrFixed', 3, ...
'ComputeKernel', 1, ...
'DataTypes', {{'MEG'}}));
% Process: Power spectrum density (Welch)
sFiles = bst_process('CallProcess', 'process_psd', sFiles, [], ...
'timewindow', timewindow.times, ...
'win_length', 1, ...
'win_overlap', 0, ...
'clusters', {}, ...
'scoutfunc', 5, ... % All
'win_std', 0, ...
'edit', struct(...
'Comment', 'Power', ...
'TimeBands', [], ...
'Freqs', [], ...
'ClusterFuncTime', 'none', ...
'Measure', 'power', ...
'Output', 'all', ...
'SaveKernel', 0));
% Process: Median: By trial group (subject average)
sFiles = bst_process('CallProcess', 'process_average', sFiles, [], ...
'avgtype', 6, ... % By trial group (subject average)
'avg_func', 8, ... % Median: median(x)
'weighted', 0, ...
'matchrows', 1, ...
'iszerobad', 1);
% Process: FOOOF: Fitting oscillations and 1/f
sFiles = bst_process('CallProcess', 'process_fooof', sFiles, [], ...
'implementation', 'matlab', ... % Matlab
'freqrange', [1, 48], ...
'peaktype', 'gaussian', ... % Gaussian
'peakwidth', [0.5, 6], ...
'maxpeaks', 6, ...
'minpeakheight', 3, ...
'proxthresh', 2, ...
'apermode', 'fixed', ... % Fixed
'guessweight', 'none', ... % None
'sorttype', 'param', ... % Peak parameters
'sortparam', 'frequency', ... % Frequency
'sortbands', {'delta', '2, 4'; 'theta', '5, 7'; 'alpha', '8, 12'; 'beta', '15, 29'; 'gamma1', '30, 59'; 'gamma2', '60, 90'});
% Process: Project on default anatomy: surface
sFiles = bst_process('CallProcess', 'process_project_sources', sFiles, [], ...
'headmodeltype', 'surface'); % Cortex surface
% Process: Spatial smoothing (10.00)
sFiles = bst_process('CallProcess', 'process_ssmooth_surfstat', sFiles, [], ...
'fwhm', 10, ...
'overwrite', 1);