Hello,
I have been given a Yokogawa MEG dataset (older NYU recordings) to analyze, and I followed your specific tutorial as well as the forum discussions about the extra head point files and Marker1 file, I still have some questions. The data do not come from the "Third party export", what I have is:
- MEG data (.sqd)
- Marker1 files (.sqd)
- .elp points from Polhemus
- .hsp points from Polhemus
-
I have a lot of headpoints per subject, which is great, but I noticed that for almost all my subjects, a lot of headpoints are outside from the helmet. Am I correct to say that it is not something I can adjust because it comes directly from the Marker and elp/hsp files? The only thing I can do is to adjust the headsurface relative to the headpoints, right?
-
Related to the previous question, if I have a really strange relationship between headpoints and helmet, it comes from my raw files and there is no way to "recover" anything for this subject?
-
What is the information read in the Marker1 file? Could you give me the link to the related brainstorm process/function?
Thank you!
Julie
Sorry for the late response: thanks, that helped a lot!!
Indeed for my question 2., the LPA and RPA were swapped, modifying the .elp file (exchanging LPA and RPA coordinates) helped put the points back on the head surface, but I still need to modify something else to put the head surface into the helmet 
it seems that the naming comes from the function you refer to (in_fopen_kit):
% === COMPUTE DIG2MEG TRANSFORMATION ===
% Compute transformation: Digitizer => MEG device
[R,T] = rot3dfit(xyzDig(:,4:end)', xyzMeg');
R = R';
T = T';
% Report in 4x4 format
header.digitize.info.digitizer2meg = [R, T; 0 0 0 1];
header.digitize.info.meg2digitizer = inv(header.digitize.info.digitizer2meg);
header.digitize.info.done = 1;
% Add the position of the digitized points to the header
nPoints = size(xyzDig,2) + size(xyzHs,2);
cellPos = num2cell([xyzDig, xyzHs]);
cellLab = cat(2, {'fidnz', 'fidt9', 'fidt10', 'LPA', 'RPA', 'CPF', 'LPF', 'RPF'}, repmat({'hs'}, 1, size(xyzHs,2)));
header.digitize.point = repmat(struct('name', '', 'x', [0 0 0], 'y', [0 0 0], 'z', [0 0 0]), [1,nPoints]);
[header.digitize.point.x] = deal(cellPos{1,:});
[header.digitize.point.y] = deal(cellPos{2,:});
[header.digitize.point.z] = deal(cellPos{3,:});
[header.digitize.point.name] = deal(cellLab{:});
But it makes me wondering what makes them different from LPA and RPA? why are these ones used to correspond to the MRI defined (blue) fiducials rather than the LPA/RPA ones?
I'm not sure I understand what you mean.
I think the MEG coils are named fid..., and then LPA etc would likely be the anatomical points. These labels seem to indicate the order in which they're expected to be digitized in this file. Sorry I don't have time right now to dig deeper into the code to fully understand.
But if you have both coils and anatomical fiducials digitized, then the latter are used to define the Brainstorm subject coordinate system (SCS) and to align with those same points picked on the MRI (prior to "refine registration with head points").
You can't directly modify the head position with respect to the helmet, that's what the head coils are for, and you have to use the "real" position. All we want to do here is make sure the MEG coils appear at their true positions with respect to the head surface. Once that's done, the position should be correct, even if it's not optimal. If it intersects with the helmet and the coils are correctly placed as best you can, then perhaps one of them actually fell off during the recording. You should be able to tell by verifying the distances between coils (from their measured positions by the MEG). Then it's more of a guessing game if you want to rescue the data.
Thanks Mark!
I think I found the problem for two of my subjects: in the marker file, either the LPA or the RPA is probably missing, so that in the function in_fopen_kit the private function getYkgwHdrCoregist sets the xyz coordinates to 0.
header.coreg = getYkgwHdrCoregist(MrkFile);
xyzMeg = cat(1, header.coreg.hpi.meg_pos)';
but I cannot open this getYkgwHdrCoregist function to further check if the Marker*.sqd file is missing these LPA/RPA coordinates or if there is some error in the getYkgwHdrCoregist function when reading the MarkerFile. I will try to place it symmetrically and I hope it will work!
Thanks!
If some anatomical fiducials were not digitized, it might be simpler to just ignore them completely, and align with the MEG head coils. In other words, whether you align with the head points or manually, you must ensure the head coils appear at the right place on the head surface, e.g. by double-checking with pictures (recommended to always take some). The location of the anatomical fiducials is not essential in comparison if you rely on the head points for the final alignment.
Hello again,
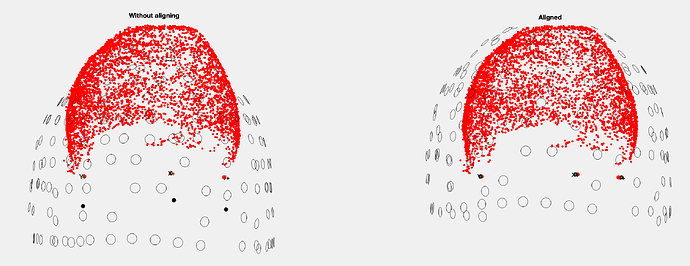
sorry to come back to you about this coregistration problem, I am still working on this project and in the meantime I have received the original old script used to process the Yokogawa MEG data in fieldtrip: In order to coregister the headpoints/yoko coils and mri fiducial points, the code in fieldtrip performs Rotation/translation transformations from the 5 coils (in the marker file) versus the digitizer (in the .elp file) and the same between the MRI fiducials to the yoko coils and digitizer fiducials, so that the visualization of the realignment looks like that:
=> before realignment the headpoints are out of the helmet and after alignment the fiducial points are very close together, which is what we want when coregistering...
I therefore looked into in_fopen_kit and everything happens very similarly with the Rotation/Translation between digitizer and MEG coils computed, etc...but somehow I am not sure about the code section called
%% ===== REGISTER WITH MRI =====
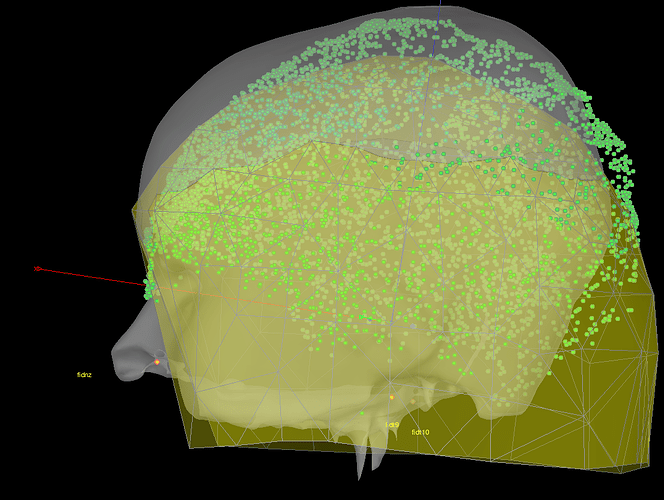
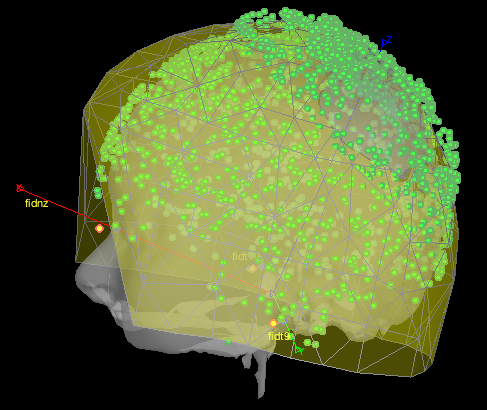
because what I then get from brainstorm is the following coregistration:
which is not satisfying and seem to be very similar to the not-aligned version of the fieldtrip plot. Is there a bug in this in_fopen_kit.m code? Or what could be the problem here?
Thanks a lot!
Sorry I don't have time to look at the code now. But from your picture, it looks like the problem is from the digitized points vs the measured positions of the head coils. It's not matching them properly, possibly matching the nasion coil (placed on the forehead perhaps?) with the digitized anatomical nasion.
Importantly, this has nothing to do with coregistration with the MRI at this stage. The relative position of head points and the helmet are independent of the MRI registration, it's aligning the digitized coils with the measured coils. So it might be they were digitized differently than expected by Brainstorm. You may want to look closely at the coordinates of each coil vs their name, and perhaps their relative distances to each other, and compare those for digitized vs measured; that might help finding the error.
Interestingly Fieldtrip doesn't seem to have that error, so it's more likely a Brainstorm issue, or FT is more robust to variations in how the coils are digitized.
Perhaps @Raymundo.Cassani or another team member can have a closer look. It would probably help if you're able to share the points and MEG files (one set) for us to test.
Thanks for your answer! How would you like me to share the files?
as far as I have looked at the code in_fopen_kit.m, until line 405 there is no apparent problem:
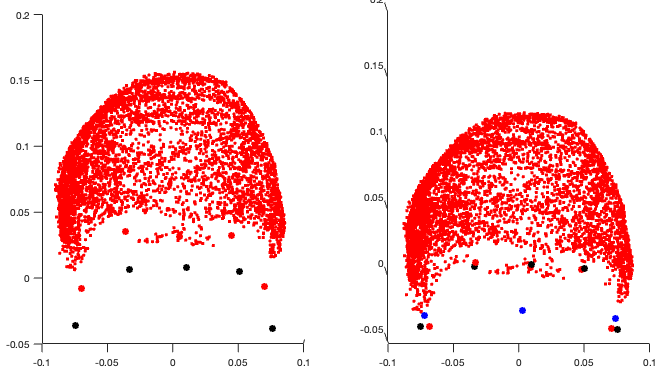
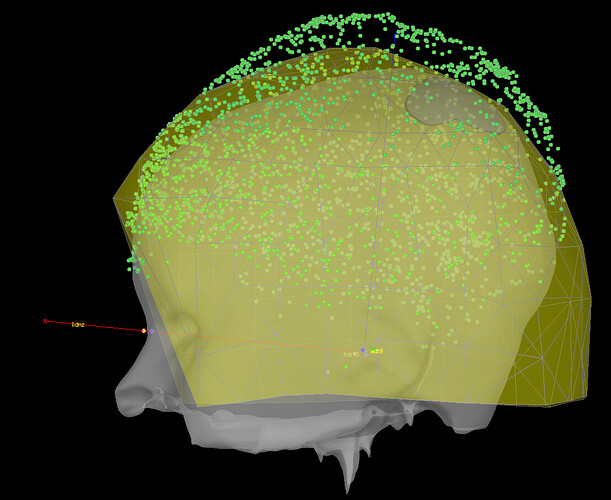
on the left I plot the xyzMeg (black), xyzDig (red) before alignment (no rotation/translation applied); on the right I plot the same xyzMeg (black), but this time I plot in red the transformed head points contained in ChannelMat.HeadPoints.Loc (with rotation and translation applied), and in blue the digitized fiducials fidz, fid9 and fid10 that will be matched to the mri fiducials later.
So it seems that ChannelMat.HeadPoints.Loc does contain headpoints coordinates that have been correctly translated 'down', so why are they out of the helmet afterwards? Are the helmet sensor information not correctly read (section from line 149)?
Regarding the following section starting line 405:
%% ===== REGISTER WITH MRI =====
it seems that cs_compute is used to get the transformation from the variable ChannelMat (containing the digitized fiducials+coils+HPI correctly transformed in MEG coordinates as seen on the plot above) to SCS, so it's not actually MRI , so you use a 'hack' because this function is supposed to be used for MRI structures in other coordinates:
% Convert the fiducials positions
% NOTE: The division/multiplication by 1000 is to compensate the T/1000 applied in the cs_convert().
% This hack was added becaue cs_convert() is intended to work on sMri structures,
% in which NAS/LPA/RPA/T fields are in millimeters, while in ChannelMat they are in meters.
might this change of units be responsible of the bug? Moreover, you therefore divide and multiply by 1000:
ChannelMat.SCS.NAS = cs_convert(ChannelMat, 'mri', 'scs', ChannelMat.SCS.NAS ./ 1000) .* 1000;
but if we have ChannelMat in meters, we should rather first multiply, to have them in millimeter for cs_convert, then divide after cs_convert, right? wouldn't something be wrong there regarding the Transfmeg etc? I have tried first multiply, then divide by 1000 and it's even worse:
the headpoints are still out of the helmet and the fiducials are not aligned at all with the MRI ones.
Please let me know how to give you the data!
Thanks for your help,
Julie
Taking a step back, I think there's no bug, but the maybe the digitized points have an issue. If you look closely at the shape of your points vs the MRI once aligned, are all the points near the head surface, and are the fiducials where you expect? Note how even in Fieldtrip they're almost touching the sensors, which means they're inside the helmet. So I don't think it's a Brainstorm issue after all.
If that's not the issue, then I had further thoughts:
The 1000 factor "hack" is done throughout Brainstorm. I don't suspect anything wrong there. And again, your problem is between the digitization and the helmet, it is independent from the MRI.
But here, you're not showing the sensors/helmet, so we don't know if the issue is present, which I presume it is. Can you add the MEG sensor locations? The helmet would be nice too, but might be a bit more complicated. If the head surface (points) is already intersecting the helmet here, then the problem could be with the detected location of those black dots vs the sensors, or possibly where Brainstorm thinks the helmet surface is vs the sensors. I'd again suggest comparing the digitized vs measured coil distances, to see if there's a notable difference, possibly indicating a coil fell off or moved. Even a small movement can cause a rotation with important effect on the other side.
If you wish to share files, please find a way for us to download them (e.g. institutional cloud or file sharing service), taking into account confidentiality issues as per your ethics guidelines of course. You can send us a private message with the info. But it doesn't look like a software issue to me anymore.
Hello Marc,
thanks for your answer!
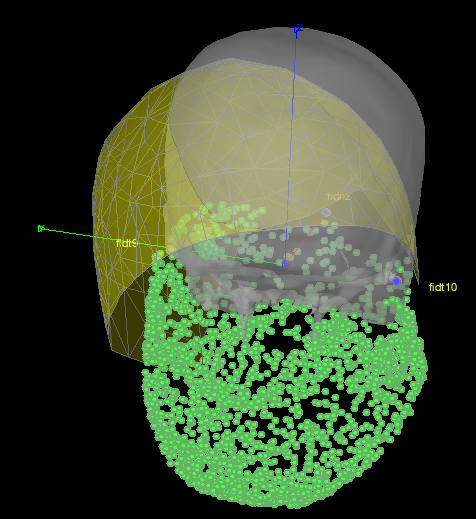
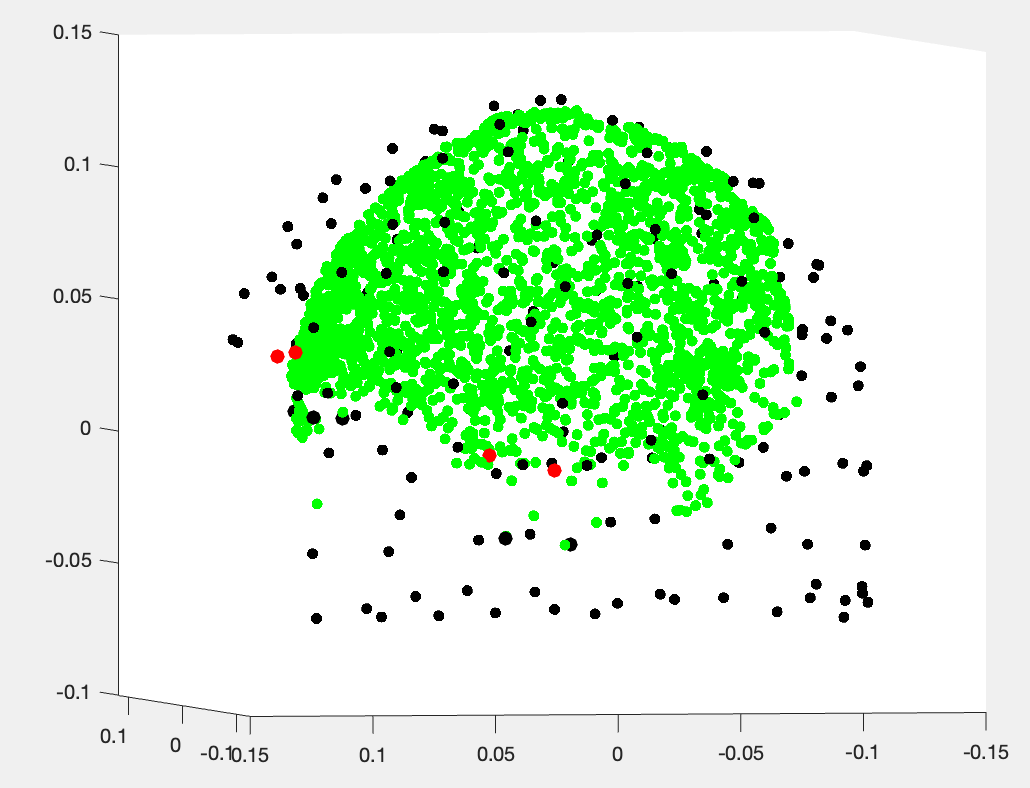
If I plot the helmet sensor position: ChannelMat.Channel(i).Loc in black (this time the headpoints are in green...):
but I might plot the wrong sensor locations? I simply plot each ChannelMat.Channel(i).Loc (x,y,z) triplets coming from:
ChannelMat.Channel(i).Loc = [header.sensors.channel(i).data.x; ...
header.sensors.channel(i).data.y; ...
header.sensors.channel(i).data.z]
all headpoints seem to be inside the helmet...
I will send the link to data through private message and further dig into that on my side!
thanks a lot,
julie
Be careful not to confuse "helmet" with "sensors". The sensors are not exactly at the surface of the helmet; there's a gap. So this figure looks like the one from Fieldtrip, which shows both software are consistent and the problem is with your data: the head is too close to the sensors and intersects the helmet surface. Sadly, we don't have the time to investigate further if the issue is not with Brainstorm but with your data. I would suggest first to try and align the MRI by marking the position of the 3 MEG head coils on the MRI (completely ignoring head points). If the head still intersects the helmet surface (not the sensors), then it means that either the head coils had moved or fallen off and were not where you thought they were, or they were not detected correctly by the MEG system. And again, you can look into the relative distances between the head coils to see if one looks like it moved. Unfortunately, I don't think there's anything more we can do.