Distortion correction using interlaced q-space sampling
Update Jan 2023: We have open-sourced the IPED code. Please see latest code at https://github.com/cbhushan/IPED. Any future updates will only be available on Github.
This page provides a precompiled Matlab implementation ("p-code") of a distortion correction method for diffusion MRI using the interlaced q-space sampling technique. This method uses interlaced phase-encoding directions (IPED) while sampling the q-space, which enables accurate distortion correction without increasing the acquisition time.
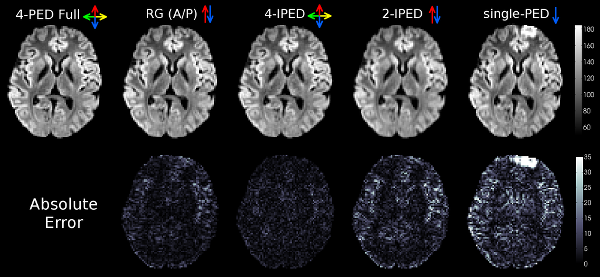 Representative comparison of applying different distortion correction methods to in-vivo data. (top) The average of all distortion-corrected DWIs from an axial slice. (bottom) Corresponding mean absolute error of the DWI image intensities, with respect to the 4-PED Full images (used as a reference since no ground truth exists for the in-vivo data). Colored arrows indicate the PED(s) used for each method. 4-PED requires four-times the regular scan time, reversed gradient (RG) method requires twice the regular scan time, while IPED and single-PED methods require regular scan time.
Representative comparison of applying different distortion correction methods to in-vivo data. (top) The average of all distortion-corrected DWIs from an axial slice. (bottom) Corresponding mean absolute error of the DWI image intensities, with respect to the 4-PED Full images (used as a reference since no ground truth exists for the in-vivo data). Colored arrows indicate the PED(s) used for each method. 4-PED requires four-times the regular scan time, reversed gradient (RG) method requires twice the regular scan time, while IPED and single-PED methods require regular scan time.
Please see following references for more details of the method:
C. Bhushan, A. A. Joshi, R. M. Leahy and J. P. Haldar (2014), Improved B0-distortion correction in diffusion MRI using interlaced q-space sampling and constrained reconstruction. Magnetic Resonance in Medicine, 72: 1218-1232. doi: 10.1002/mrm.25026
C. Bhushan, A. A. Joshi, R. M. Leahy and J. P. Haldar (2013), Accelerating data acquisition for reversed-gradient distortion correction in diffusion MRI: A constrained reconstruction approach. 21st Scientific Meeting of International Society for Magnetic Resonance in Medicine (ISMRM), Salt Lake City, p. 55. Abstract PDF, ISMRM presentation video
Data acquisition
 This implementation makes several assumptions about the data acquisition, data format and sampling pattern in the q-space. All of the following must be satisfied for accurate correction:
This implementation makes several assumptions about the data acquisition, data format and sampling pattern in the q-space. All of the following must be satisfied for accurate correction:
- The diffusion data should be scanned with single-shot Echo Planar Imaging (EPI) sequence.
- The EPI planes for data acquired with different phase encoding directions (PED) must coincide. This also implies that ignoring the polarity of PEDs there could be maximum of two PEDs, one along each dimension of 2D EPI plane (without ignoring the polarity of PEDs there could be maximum of four PEDs).
- Atleast two different PEDs must be used to sample the q-space.
All the diffusion weighted images must be acquired with a single b-value i.e. assumes single-shell acquisition (multi-shell acquisition can also be used - by correcting diffusion data for each shell separately).
- Atleast one diffusion image must be acquired with (almost) no diffusion weighting (b-value of zero) for each unique PED.
The inhomogeneity in main magnetic field (B0) should be same across all diffusion weighted images i.e. all the diffusion images must be acquired with same shim settings.
- The field of view, voxel resolution, image matrix size, number of slices and slice thickness should be consistent across all images.
- All the input diffusion data should be the reconstructed magnitude diffusion weighted images. Complex k-space data (or raw-data) is not supported in this implementation.
Further, take note about following to enable accurate distortion correction with IPED method with this implementation:
- It is recommended to use more than 20 unique diffusion encoding directions. Larger number of diffusion encoding direction would enable better correction.
- The diffusion encoding directions should be evenly distributed on the sphere (or one hemisphere). The sampling directions are plotted for easy visualization in this implementation.
The diffusion encoding directions for each PEDs should be evenly interlaced i.e. encoding directions should be evenly distributed for each unique PED as well as for union of all PEDs.
- In case of multi-shell acquisition, the above recommendation applies to each shell.
For data acquired using parallel imaging technique, the input echo spacing should be set to the effective echo spacing. Eg. if a 2X acceleration was achieved with parallel imaging then the effective echo spacing should be half of the echo spacing used in the sequence.
This implementation also works with reversed-gradient (Chang and Fitzpatrick, 1992) and its generalized acquisition schemes without any modification. Also see optional inputs below for more options about generalized reversed-gradient schemes.
Usage
The distortion correction method is implemented as a Matlab function, named IPEDcorrect, and requires Image processing toolbox. It only supports use of NIfTI-1 data format for image and plain text-files for specifying other parameters. Either of following syntax is supported:
IPEDcorrect(IPED_config_filename, fieldmap_filename, echo_spacing, output_base) IPEDcorrect(IPED_config_filename, fieldmap_filename, echo_spacing, output_base, opts)
where,
IPED_config_filename is a string specifying the file-name of the IPED-config file. See below for more details about format of the IPED-config file.
fieldmap_filename is a string specifying the file-name of the fieldmap saved in NIfTI-1 format. The fieldmap can acquired or estimated using any method but it must be specified in units of radians/sec. The voxel resolution of fieldmap need not match to that of the diffusion images, however for best correction it should cover the field-of-view of the diffusion scans.
echo_spacing is a number specifying the echo-spacing in units of secs. Eg: For an echo spacing of 0.36ms, use 0.00036.
output_base is a string specifying the file-prefix for the outputs written by IPEDcorrect. Eg: when output_base is set to sub02.IPED-corrected, the file-names of the outputs written by IPEDcorrect would be sub02.IPED-corrected.nii.gz, sub02.IPED-corrected.bvec and likewise.
opts is an optional input which specifies several optional arguments, like regularization parameters, for distortion correction. See below for more details.
IPED-config file
IPED-config file is a plain text file which describes different acquisition parameters for the diffusion data. The file follows a table-type format, which can be created and saved using a simple text editor. The IPED-config file can be saved with any extension. Each row of the file describes parameter for diffusion images corresponding to one phase-encoding direction. Number of columns in each row should be consistent and must have three, four or five columns. The first column specifies the filename of the 4D NIfTI-1 diffusion file. Second column specifies the phase encoding direction (PED). Rest of the columns specifies filenames which describe diffusion encoding parameters as shown below. In addition, any text coming after a # (hash) is ignored for rest of the line.
Four columns (with bvec, bval file):
4D-nifti-filename.nii.gz PED bvec-filename bval-filename
Three columns (with bmat file):
4D-nifti-filename.nii.gz PED bmat-filename
Five columns (with bvec, bval and bmat file):
4D-nifti-filename.nii.gz PED bvec-filename bval-filename bmat-filename
Filenames can be specified either with their full-path or without any path (for the later case, files are assumed to be in the same directory as the IPED-config file. Relative paths (eg: subdirectory/file.nii.gz) are not supported. When the filenames/paths contain spaces then they must be enclosed in double quotes (eg: "subject23 trial2.nii.gz"). Different columns of the config file can also be delimited by comma, like that in standard csv files (in case of csv format, filenames with spaces can be directly specified and must not use double quotes).
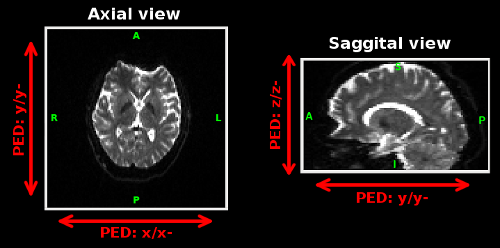 EPI images are most distorted along the phase encoding direction (PED) and IPEDcorrect formulates the distortion correction as a 1D problem. PED can be easily be specified after visualizing the images in BrainSuite software. Valid options for PED are x, x-, y, y-, z and z-. x direction increases towards the right side of the subject, while x- increases towards the left side of the subject. Similarly, y and y- are along the anterior-posterior direction of the subject, and z and z- are along the inferior-superior direction. The figure on the right describes the PED directions in different views. The image shown in the above figure (full BrainSuite view) has PED of y-. The polarity of the PED depends on the sign convention of the fieldmap and it can be best adjusted by trying the function with different polarity.
EPI images are most distorted along the phase encoding direction (PED) and IPEDcorrect formulates the distortion correction as a 1D problem. PED can be easily be specified after visualizing the images in BrainSuite software. Valid options for PED are x, x-, y, y-, z and z-. x direction increases towards the right side of the subject, while x- increases towards the left side of the subject. Similarly, y and y- are along the anterior-posterior direction of the subject, and z and z- are along the inferior-superior direction. The figure on the right describes the PED directions in different views. The image shown in the above figure (full BrainSuite view) has PED of y-. The polarity of the PED depends on the sign convention of the fieldmap and it can be best adjusted by trying the function with different polarity.
The bvec and/or bmat files should describe diffusion encoding direction in voxel coordinate of the diffusion data (i.e. in the same frame of reference as the image matrix). Further, the order of entries of gradient direction in the specified .bvec or .bmat file must match the order of volumes in the input 4-D NIfTI data.
Following is an example of the content of an IPED-config file with four different PEDs.
6067JH.DWI.A.nii.gz y- 6067JH.DWI.A.bvec 6067JH.DWI.A.bval 6067JH.DWI.P.nii.gz y 6067JH.DWI.P.bvec 6067JH.DWI.P.bval 6067JH.DWI.L.nii.gz x- 6067JH.DWI.L.bvec 6067JH.DWI.L.bval 6067JH.DWI.R.nii.gz x 6067JH.DWI.R.bvec 6067JH.DWI.R.bval
Optional inputs
IPEDcorrect can take several optional inputs to change the behavior of the different regularization terms.
opts.spatial_beta: Specifies the weight for spatial regularization.
opts.spherical_alpha: Specifies the weight for spherical regularization.
opts.shOrder: Maximum order of the spherical-harmonic basis function used for correction. Must be even integer greater than zero.
opts.sph_wt: Specifies the nature of the voxel-wise weight for spherical regularization. It can take three possible values:
When opts.sph_wt is empty string then same weight (opts.spherical_alpha) is used for all voxels.
When opts.sph_wt is set to 'G_mag' then the spherical weight at each voxel is scaled by magnitude of the gradient of the fieldmap at each voxel. In this case, opts.G_mag_low and opts.G_mag_high is further used to threshold the magnitude of the gradient (in units of rad/sec/mm) as described in the paper.
opts.sph_wt can be set to name of a 3D NIfTI file, values of which is then used to scale the spherical weight at each voxel.
opts.estimation_mask: Specifies the filename of a mask. When specified, this mask is used the specify the region in which the diffusion images are corrected for distortion. Usually, use of a mask has substantial impact on the runtime.
opts.save_SH_coeff: Specifies how to save the estimated spherical-harmonic (SH) coefficients for diffusion weighted images. It can take three numerical values:
0 : Do not save SH-coefficients.
1 : Save BrainSuite style SH-coefficients (3D NIfTI volumes for each coefficient in a folder named DWI_IPED_SHcoeff.
2 : Save a 4D NIfTI volume of SH-coefficients.
opts.isfull: Specifies if each of the diffusion encoded image is acquired with all the PEDs. This flag only affects the format of the final output and not the estimates themselves. When set to true, each of the PEDs must have same number of diffusion encoding and same number of volumes in 4D NIfTI files.
Following Matlab structure defines the default options used in IPEDcorrect:
opts = struct(... 'spatial_beta', 0.27, ... 'spherical_alpha', 0.04, ... 'shOrder', 8, ... 'sph_wt', 'G_mag', ... 'G_mag_low', 1*2*pi, ... 'G_mag_high', 9*2*pi, ... 'estimation_mask', '', ... 'isfull', false, ... 'save_SH_coeff', 1 ... );
Download & Installation
This code is licensed under the CC Attribution-Noncommercial-Share Alike 4.0 License. Please cite [1] and [2] if you use this code or its derivatives in your own work.
In no event shall the University of Southern California, the Authors, or the Distributors be liable to any party for direct, indirect, special, incidental, or consequential damages, including lost profits, arising out of the use of this software, its documentation, or any derivatives thereof, even if the authors have been advised of the possibility of such damage. The University of Southern California, the Authors, and the Distributors specifically disclaim any warranties, including, but not limited to, the implied warranties of merchantability, fitness for a particular purpose, and non-infringement. This software is provided on an "as is" basis, and the authors and distributors have no obligation to provide maintenance, support, updates, enhancements, or modifications. This software is for research purposes only and has not been approved for clinical use.
Download IPEDcorrect_v19.zip (Last Modified: Feb 12, 2017)
System requirements
IPEDcorrect is a Matlab function and requires Image processing toolbox. Only 64-bit platforms (Windows, MacOS, Linux) are supported. It has been only tested with Matlab 2012a and Matlab 2014b. Memory requirement depends of size of the input diffusion dataset. opts.estimation_mask can be used to substantially lower the memory requirement and run-time.
Installation
Download the zip file and extract the content to a directory on your computer, say /home/username/matlab/IPED.
Add the directory to Matlab path by using following command on Matlab prompt (appropriately replace the name of directory): addpath(genpath('/home/username/matlab/IPED'))
IPEDcorrect should be available in Matlab after adding path. Test it by typing IPEDcorrect on Matlab command prompt. It should issue an error about missing input arguments, as shown below.
>> addpath(genpath('/home/username/matlab/IPED'))
>> IPEDcorrect
Error using IPEDcorrect (line 12)
Incorrect number of input arguments. IPEDcorrect() needs either 4 or 5 input
arguments. Please see usage details at:
http://neuroimage.usc.edu/neuro/Resources/IPED