Export source maps to SPM8
The statistical analysis is still limited in Brainstorm, but you can easily export your source maps and run your tests with an external application. This tutorial explains how to export source maps to SPM8. It is based on the median nerve tutorial dataset that was used in the tutorials of the section ?Processing continuous recordings. If you have followed those tutorials, this dataset should be available in your database in the protocol TutorialRaw.
SPM8 has an important limitation with respect with SPM12: it cannot work on surfaces. We have to export all the sources to full volumes, which is highly inefficient. If you get the option, consider updating to SPM12.
Install SPM8
Installing SPM is easy: http://www.fil.ion.ucl.ac.uk/spm/software/spm8/
Fill up the registration form, download the file "spm8.zip" and unzip it.
Calculate unconstrained sources
In the previous tutorial, we have calculated sources constrained to the cortex surface. Because SPM8 works only with full volumes, we are going to estimate the sources in the full head volume, as detailed in the tutorial Volume source estimation.
Right-click on (common files) > Compute head model > MRI volume.
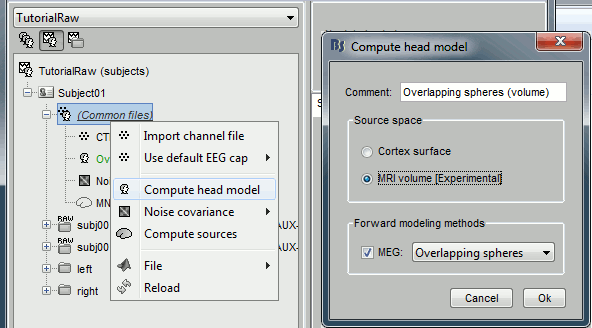
Right-click on the new head model > Compute sources > keep all the default options.
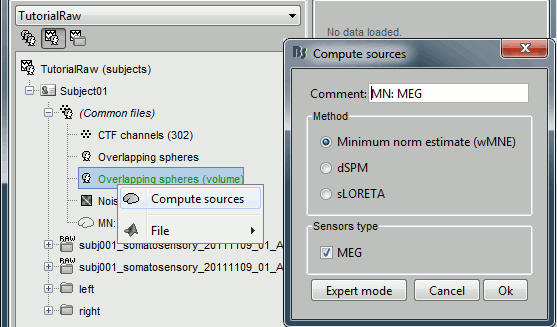
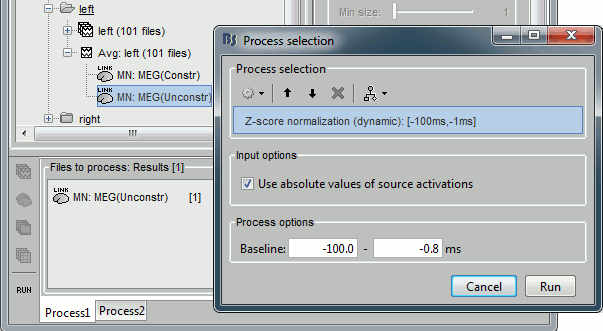
Pick any of the unconstrained source file and drop it in the Process1 box. Run the process "Standardize > Z-score (dynamic)". It creates a new shared source file, available for all the files of Subject01.
Export source files
We want now to export the z-scored unconstrained sources to volume files in NIfTI1 format (readable by SPM). We are going to get rid of the time information, very difficult to handle with those full volumes in SPM, and keep only two averaged time windows per condition: a baseline [-20,-15]ms and an active state [33,38]ms. We are going to generate 2*101 left + 2*98 right .nii files.
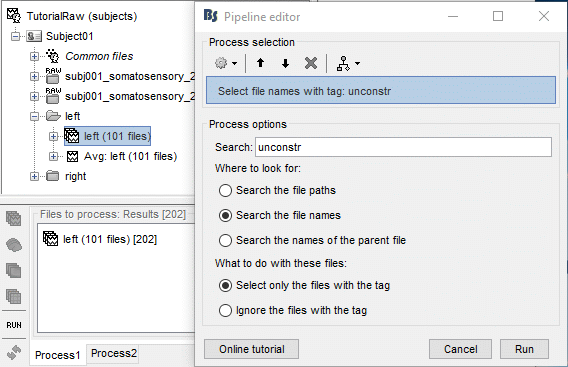
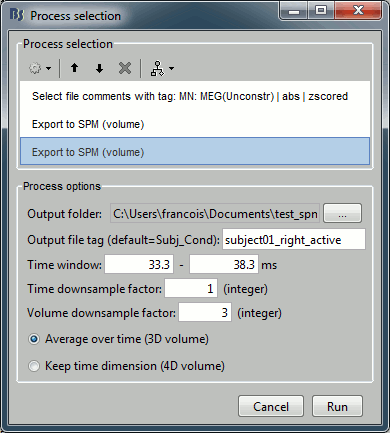
Clear the Process1 box and drop all the trials for condition Left in it. Select the button "Process sources" on the left of the Process1 tab. To select only the unconstrained/z-scored files, you could simply use the checkbox "zscore", but we are going to illustrate a more generic solution: select the process "File > Select files with tag", illustrated in the tutorial Selecting files in the database. Configure it as following:
Then in the same pipeline, select the process "File > Export to SPM".
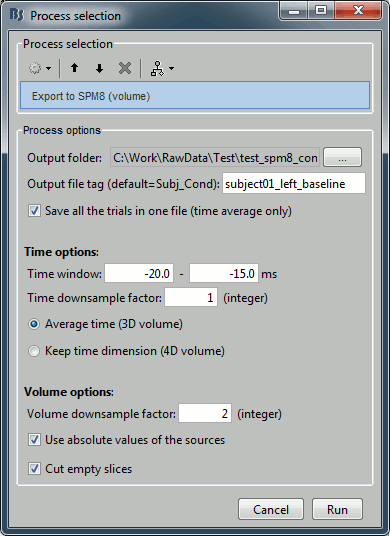
Configure the process:
Output folder: Select the folder where you want all the .nii files to be saved
Output file tag: Set the file name to this exported .nii files. If you leave this option empty, the default file names would be "Subject01_Left_*.nii", which is not informative enough, given that we want to export two states for the same condition: baseline vs. active.
Time window: Segment of the file that we want to export. We start with the baseline: -20ms to -15ms
Time downsample factor: If we keep the time information, we can save only one time sample every N in the .nii files, to limit the size of the generated files. The value we set here is not used because we specify in the last option that we average in the time dimension.
Volume downsample: Set it to 3, it will take one voxel every three voxels in each dimension. The MRI volume we imported in the Brainstorm database is 256x256x256, the output volumes with the functional data are only 86x86x86, again to limit the size of the files we generate at this step.
Average over time / Keep time dimension: Defines what we do with the time. In this case, we want to average over the small time windows to get rid of the time.
Add another export process right after this one, to export the "active" state. Set the time window to [33,38]ms, to capture the peak of the response in the primary somatosensory cortex, and the file name to "subject01_left_active".
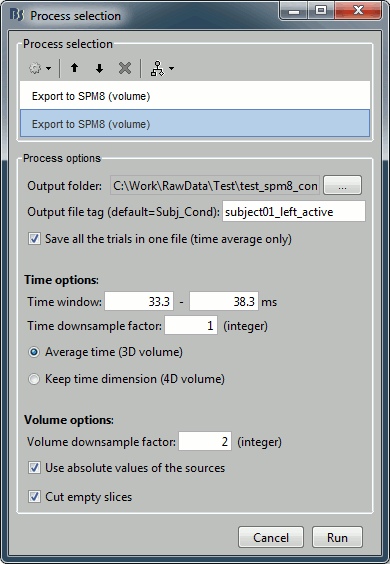
Click on Run and let it process for a while. It will generate 2*101=202 NIfTI files in the output folder.
Repeat the operation for the right condition: change only the file output names to "subject01_right_baseline" and "subject01_right_active".
Open you file explorer and check the .nii files that were created in the destination folder.

You can try open some of those files with a NIfTI file viewer, just to make sure that what is saved is coherent. You can use for instance the free software Mricron to view the file subject01_right_active_02.nii. This a stimulation of the right median nerve, we can see clearly that the response is located in the central regions of the left hemisphere, which is what we expect.
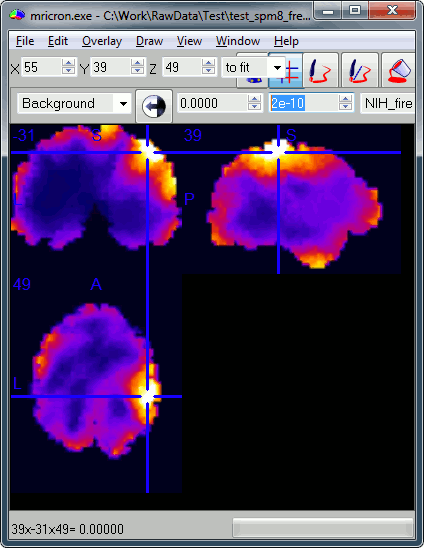
Note that you can also you Brainstorm as a NIfTI viewer: go to the anatomy view of the protocol, right-click on Subject01 > Import MRI > Select a .nii file.
Start SPM
Start Matlab, go to the SPM folder, add it to your path if it is not done already, and type "spm".