|
Size: 11473
Comment:
|
Size: 14766
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 6: | Line 6: |
| <<TableOfContents(2,2)>> | <<TableOfContents(3,2)>> |
| Line 9: | Line 9: |
| PLS is a free toolbox that is available at Baycrest (https://www.rotman-baycrest.on.ca/index.php?section=84). The PLS code is written entirely in MATLAB (Mathworks Inc) and can be downloaded from https://www.rotman-baycrest.on.ca/index.php?section=345. To cite PLS Toolbox, please see the “References” section of this tutorial. | PLS is a free toolbox that is a part of [[https://www.rotman-baycrest.on.ca/index.php?section=84|Baycrest, University of Toronto]]. The PLS code is written entirely in MATLAB (Mathworks Inc) and can be downloaded from https://www.rotman-baycrest.on.ca/index.php?section=345. To cite PLS Toolbox, please see the “References” section of this tutorial. |
| Line 12: | Line 12: |
| PLS analysis is explained in general in this section. However, this tutorial assumes that the users are already familiar with basics of PLS analysis and it does not include details about this technique. If PLS is new to you or if you want to read more about PLS and its applications in details, please refer to “References” and relevant articles introduced here. | Partial Least Squares (PLS) analysis is a multivariate statistical technique that is used to find the relationship between two blocks of variables. PLS that has various applications and types (Krishnan et al., 2011); however, the focus of this tutorial is on Mean-Centered PLS analysis, which is a common type of PLS while working with neuroimaging data. In this type of PLS analysis, one data block is neural activity (e.g. MEG measurements/source data here) while the other one is the experiment design (e.g. different groups/conditions). |
| Line 14: | Line 14: |
| Partial Least Squares (PLS) analysis is a multivariate statistical technique that is used to find the relationship between two blocks of variables. PLS that has various applications and types (Krishnan et al); however, the focus of this tutorial is on Mean-Centered PLS analysis, which is a common type of PLS while working with neuroimaging data. In this type of PLS analysis, one data block is neural activity (e.g. MEG measurements/source data here) while the other one is the experiment design (e.g. different groups/conditions). | PLS analysis is based on extracting the common information between the two data blocks by finding a correlation matrix and linear combinations of variables in both data blocks that have maximum covariance with one another. In the example provided here, we find a contrast between different conditions as well as patterns of brain activity that maximally covary with that specific contrast. |
| Line 16: | Line 16: |
| PLS analysis is based on extracting the common information between the two data blocks by finding a correlation matrix and linear combinations of variables in both data blocks that have maximum covariance with one another. In the example provided here, we find a contrast (or an effect) between different conditions as well as patterns of brain activity that maximally covary with that specific contrast. | For this purpose, we take the neural activity as one data block, matrix X, where the rows of matrix X are observations (participants/trials) nested in conditions or groups, and the columns of X are variables that are arranged in a way that time scales are nested within sources. The other data block, matrix Y, is a matrix of dummy coding that is related to experimental design (different groups or conditions) (Krishnan et al., 2011). |
| Line 18: | Line 18: |
| For this purpose, we take the neural activity as one data block, matrix X, where the rows of matrix X are observations (participants/trials) nested in conditions or groups, and the columns of X are variables that are arranged in a way that time scales are nested within sources. The other data block, matrix Y, is a matrix of dummy coding that is related to experimental design (different groups or conditions) (Krishnan et al). | PLS analysis first calculates a mean-centered matrix using matrices X and Y. Then, singular value decomposition (SVD) is applied on the mean-centered matrix. The outcome of PLS analysis is a set of latent variables that are in fact linear combinations of initial variables of the two data blocks that maximally covary with the resulting contrasts (Krishnan et al., 2011, Mišić et al., 2016). |
| Line 20: | Line 20: |
| PLS analysis first calculates a mean-centered matrix using matrices X and Y. Then, singular value decomposition (SVD) is applied on the mean-centered matrix. The outcome of PLS analysis is a set of latent variables that are in fact linear combinations of initial variables of the two data blocks that maximally covary with the resulting contrasts (419, Krishnan et al., 2011). Finally, the statistical significance of a latent variable is defined by a p-value from permutation testing. In addition, bootstrapping is used to assess the reliability of each original variable (e.g. a source at a time point) that contributes to the latent variable. Bootstrap ratios are calculated for each original variable for this purpose. More specifically, each latent variable consists of a set of singular values that describe the effect size, as well as a set of singular vectors, ''or weights'', that define the contribution of each initial variable to the latent variables. The ratio of these weights to the standard errors estimated from bootstrapping is called bootstrap ratio. Therefore, the larger the magnitude of a bootstrap ratio, the larger the weight (i.e. contribution to the latent variable) and the smaller the standard error (i.e. higher stability) (McIntosh and Lobaugh 2004, 419). Bootstrap ratio can be equivalent to a z-score if we have an approximately normal bootstrap distribution (Efron and Tibshirani, 1986). |
Finally, the statistical significance of a latent variable is defined by a p-value calculated from permutation test. In addition, bootstrapping is used to assess the reliability of each original variable (e.g. a source at a time point) that contributes to the latent variable. Bootstrap ratios are calculated for each original variable for this purpose. More specifically, each latent variable consists of a set of singular values that describe the effect size, as well as a set of singular vectors, ''or weights'', that define the contribution of each initial variable to the latent variables. The ratio of these weights to the standard errors estimated from bootstrapping is called bootstrap ratio. Therefore, the larger the magnitude of a bootstrap ratio, the larger the weight (i.e. contribution to the latent variable) and the smaller the standard error (i.e. higher stability) (McIntosh and Lobaugh, 2004, Mišić et al., 2016). Bootstrap ratio can be equivalent to a z-score if we have an approximately normal bootstrap distribution (Efron and Tibshirani, 1986). |
| Line 30: | Line 28: |
| * * Decoding > Classification with cross-validation (process_decoding_crossval.m) * Decoding > Classification with permutation (process_decoding_permutation.m) |
The data processed here is the same dataset that is used in [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingle|MEG visual tutorial: Single subject]] and [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup|MEG visual tutorial: Group analysis]]'''. '''This dataset consists in simultaneous MEG/EEG recordings of 19 subjects performing a simple visual task on a large number of famous, unfamiliar and scrambled faces. The detailed presentation of experiment is available in the [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingle|MEG visual tutorial: Single Subject]]. |
| Line 34: | Line 30: |
| These two processes work in a similar way: | You can follow this tutorial after processing the data as illustrated in [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingle|MEG visual tutorial: Single Subject]].''' '''Then: |
| Line 36: | Line 32: |
| * '''Input''': the input is the channel data from two conditions (e.g. condA and condB) across time. Number of samples per condition should be the same for both condA and condB. Each of them should at least contain two samples. * '''Output''': the output is a decoding curve across time, showing your decoding accuracy (decoding condA vs. condB) at time point 't'. * '''Classifier''': Two methods are offered for the classification of MEG recordings across time: Support vector machine (SVM) and Linear discriminant analysis (LDA). |
* Find the subject averages as it is explained in [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup|MEG visual tutorial: Group analysis]] by following the steps in the visual group analysis up to [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Subject_averages:_Filter_and_normalize|Section 7: Subject averages: Filter and normalize]].''' ''' * After you found all the averages across subjects, continue with [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Subject_averages:_Filter_and_normalize|Section 7]] and filter the signals below 32Hz and extract time as it is explained. However, [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Sources-2|when filtering the sources]], __do not__ normalize the __source__ values with respect to baseline (i.e. __do not__ find z-score). * At this point, continue with the rest of the group analysis tutorial: [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Projet_sources_on_template|Project sources on template]] and [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Spatial_smoothing|apply spatial smoothing on the source maps]]. * Data is now ready for PLS analysis. |
| Line 40: | Line 37: |
| In the context of this tutorial, we have two condition types: faces, and scenes. We want to decode faces vs. scenes using 306 MEG channels. In the data, the faces are named as condition ‘201’; and the scenes are named as condition ‘203’. | == Running PLS == There are two PLS processes available in Brainstorm: * You can run PLS analysis for __only two conditions__ through the Process2 tab at the bottom of the Brainstorm window. |
| Line 42: | Line 40: |
| * You can also run PLS analysis for __more than two conditions__ through the Process1 tab at the bottom of the Brainstorm window. This option is explained in the “Advanced” section of this tutorial. <<BR>> | |
| Line 43: | Line 42: |
| Both of these two processes work in a similar way: | |
| Line 44: | Line 44: |
| * Go to the [[http://neuroimage.usc.edu/bst/download.php|Download]] page of this website, and download the file: '''sample_decoding.zip ''' * Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder). This is really important that you always keep your original data files in a separate folder: the program folder can be deleted when updating the software, and the contents of the database folder is supposed to be manipulated only by the program itself. |
* '''Input: '''the input files are the source data from different conditions. Number of samples per condition must be the same for all conditions. Make sure you have enough observations/trials per condition in order to get meaningful results from statistical tests. * '''Note: You can also run PLS analysis on sensor level data (channel data). However, the results will not be as meaningful and useful as the ones from source level data.''' * '''Output: ''' the output files include bootstrap ratios and p-values for all latent variables. In addition, you can look at the contrast between conditions (groups and/or experimental tasks) for each latent variable. The output files are explained in details in the following sections.<<BR>> |
| Line 47: | Line 48: |
| * Start Brainstorm (Matlab scripts or stand-alone version). * Select the menu File > Create new protocol. Name it "'''TutorialDecoding'''" and select the options: * "'''Yes, use protocol's default anatomy'''", * "'''No, use one channel file per condition'''". <<BR>><<BR>> {{attachment:decoding_protocol.gif||height="370",width="344"}} |
We will continue with explaining the PLS process for only two conditions from now and we will leave PLS process for more conditions for the Advanced section of this tutorial. |
| Line 52: | Line 50: |
| == Import the recordings == * Go to the "functional data" view (sorted by subjects). * Right-click on the TutorialDecoding folder > New subject > '''Subject01''' <<BR>>Leave the default options you defined for the protocol. * Right click on the subject node (Subject01) > '''Review raw file'''''.'' <<BR>>Select the file format: "'''MEG/EEG: Neuromag FIFF (*.fif)'''"<<BR>>Select the file: sample_decoding/'''mem6-0_tsss_mc.fif''' <<BR>><<BR>> {{attachment:decoding_link.gif||height="169",width="547"}} * Select "Event channels" to read the triggers from the stimulus channel. <<BR>><<BR>> {{attachment:decoding_events.gif||height="162",width="289"}} * We will not pay much attention to MEG/MRI registration because we are not going to compute any source models, the decoding is done on the sensor data. * Right-click on the "Link to raw file" > '''Import in database'''. <<BR>><<BR>> {{attachment:decoding_import.gif||height="317",width="571"}} * Select only two events: 201 (faces) and 203(scenes) * Epoch time: [-100, 1000] ms * Remove DC offset: Time range: [-100, 0] ms * Do not create separate folders for each event type * You will get a message saying "some epochs are shorter than the others". Answer '''yes'''. |
=== Select files === * Select the process2 tab at the bottom of Brainstorm window. * Drag and drop '''16 source files''' from '''Group_analysis/Faces-MEG''' to the left (Files A). * Drag and drop '''16 source files''' from '''Group_analysis/Scrambled-MEG''' to the right (Files B). * Note that the number of files in each window (“A” and “B”) must be the same.<<BR>> <<BR>> {{attachment:select_files.gif}} |
| Line 65: | Line 56: |
| == Select files == Select the Process2 tab at the bottom of the Brainstorm window. |
=== Run PLS === * In Process2, click on [Run]. * Select process '''Test > Partial Lest Squares (PLS). '''This opens the '''Pipeline editor''' window with the PLS process for two conditions.<<BR>><<BR>> {{attachment:pipeline1.gif}} |
| Line 68: | Line 60: |
| * Drag and drop '''40 files''' from group '''201''' to the left (Files A). * Drag and drop '''40 files''' from group '''203''' to the right (Files B). * You can select more than 40 or less. The important thing is that both ‘A’ and ‘B’ should have the same number of files. <<BR>><<BR>> {{attachment:decoding_selectfiles.gif||height="369",width="398"}} |
* '''“Condition 1” and “Condition 2”''': Name of the two input conditions: “Condition 1” and “Condition 2” are related to files in windows “A” and “B” of “Process2” tab, respectively. We can use “Faces” and “Scrambled Faces” as the condition names here. |
| Line 72: | Line 62: |
| == Cross-validation == '''Cross'''-'''validation''' is a model '''validation''' technique for assessing how the results of our decoding analysis will generalize to an independent data set. |
* '''Number of Permutations''': Indicates the number of permutations you want to run in PLS. * '''Number of Bootstraps''': Indicates the number of bootstraps you want to run in PLS. * '''Sensor types or name''': Indicates the type of sensors that has been used for source localization (e.g. MEG or EEG). This will occur in the result file name as well. |
| Line 75: | Line 66: |
| * Select process "'''Decoding > Classification with cross-validation'''": <<BR>><<BR>> {{attachment:cv_process.gif||height="388",width="346"}} * '''Low-pass cutoff frequency''': If set, it will apply a low-pass filter to all the input recordings. * '''Matlab SVM/LDA''': Require Matlab's Statistics and Machine Learning Toolbox. <<BR>>These methods do a k-fold stratified cross-validation for you, meaning that each fold will contain the same proportions of the two types of class labels (option "Number of folds"). * '''LibSVM''': Requires the LibSVM toolbox ([[http://www.csie.ntu.edu.tw/~cjlin/libsvm/#download|download]] and add to your path).<<BR>>The LibSVM cross-validation may be faster but will not be stratified. |
Run the process when the options are set. The process will take some time depending on the number of files, number of permutations and bootstraps. The results are then saved in '''Group Analysis > Intra-Subject''' folder. |
| Line 80: | Line 68: |
| * The process will take some time. The results are then saved in a file in the new decoding folder. <<BR>><<BR>> {{attachment:cv_file.gif||height="167",width="268"}} * If you double click on it you will see a decoding curve across time. <<BR>><<BR>> {{attachment:cv_plot.gif||height="172",width="355"}} |
=== PLS Results === * Output files include contrast between conditions, p-value of the latent variable and bootstrap ratios:<<BR>><<BR>> {{attachment:results_overview.gif}} |
| Line 83: | Line 71: |
| == Permutation == This is an iterative procedure. The training and test data for the SVM/LDA classifier are selected in each iteration by randomly permuting the samples and grouping them into bins of size n (you can select the trial bin sizes). In each iteration two samples (one from each condition) are left out for test. The rest of the data are used to train the classifier with. |
<<BR>> |
| Line 86: | Line 73: |
| * Select process "'''Decoding > Classification with cross-validation'''". Set options as below: <<BR>><<BR>> {{attachment:perm_process.gif||height="366",width="311"}} * '''Trial bin size''': If greater than 1, the training data will be randomly grouped into bins of the size you determine here. The samples within each bin are then averaged (we refer to this as sub-averaging); the classifier is then trained using the averaged samples. For example, if you have 40 faces and 40 scenes, and you set the trial bin size to 5; then for each condition you will have 8 bins each containing 5 samples. Seven bins from each condition will be used for training, and the two left-out bins (one face bin, one scene bin) will be used for testing the classifier performance. * The results are saved in a file in the new decoding folder. <<BR>><<BR>> {{attachment:perm_file.gif||height="142",width="284"}} * Right-click > Display as time series (or double click). <<BR>><<BR>> {{attachment:perm_plot.gif||height="169",width="346"}} |
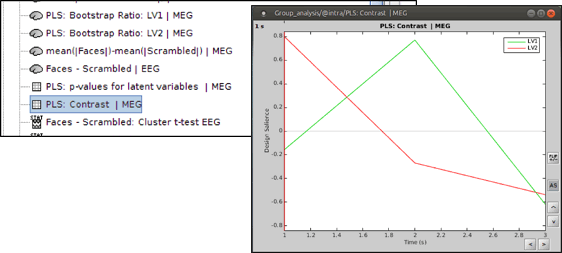
* '''PLS : Contrast: '''You can first look at the contrast between two conditions: Double-click on the contrast file. The contrast is shown in the form of time series; however, the x-axis is not actual time. In fact, the integer numbers show the number of conditions (e.g. 1 for condition 1 or “Faces” and 2 for Condition 2 or “Scrambled Faces”) and the non-integer ones should be ignored.<<BR>><<BR>> {{attachment:contrast.gif}} |
| Line 91: | Line 75: |
| == Acknowledgment == This work was supported by the ''McGovern Institute Neurotechnology Program'' to PIs: Aude Oliva and Dimitrios Pantazis. ''http://mcgovern.mit.edu/technology/neurotechnology-program'' |
<<BR>> * '''PLS: p-value for latent variable:''' if you double click on this files, you will see a table containing the p-value of the latent variable that is related to the contrast shown above. A significant latent variable means the contrast observed between two conditions is also significant. * Note: You will see two numbers in the table for p-value of the latent variable when you run PLS for two conditions. Ignore the second one.<<BR>><<BR>> {{attachment:p_value.gif}} <<BR>> * '''PLS: Bootstrap ratio: '''This file is saved in the format of source maps. Double-click on open the file. The colormap shows bootstrap ratios for each source at each time point. There are some important points you should keep in mind when working with bootstrap ratios: * Bootstrap ratios show how reliably each source is contributing to the observed contrast and latent variable at each time point. A bootstrap ratio larger than 2.58 is considered reliable. Therefore, adjust the amplitude of bootstrap ratios to 25%-26% to be able to see the reliable signals. For this, choose “Surface” tab from Brainstorm window and adjust the “Amplitude”.<<BR>><<BR>> {{attachment:bootstrap_ratio.gif}} * Do not display absolute values of the signals. Make sure “Absolute Values” is not selected in the colormap menu for this figure. The sign of bootstrap ratio is important since positive bootstrap ratios express the contrast as is, while bootstrap ratios with negative values express the opposite contrast. * You can display the results at different time points using contact sheet. <<BR>><<BR>> {{attachment:contact_sheet.gif}} <<BR>> * You can also define regions of interest as usual: OFA (Occipital Face Area), V1.<<BR>><<BR>> {{attachment:ROI.gif}} <<TAG(Advanced)>> == Advanced Option == You can run PLS for more than 2 conditions through Process1 tab. However, you should be careful when arranging the files in this option. We will use the same dataset to illustrate how to use this option in Brainstorm. * Select the Process1 tab at the bottom of Brainstorm window. * Drag and drop '''16 source maps''' from '''Group_analysis/Famous-MEG''' to the Process1 window. * Drag and drop '''16 source maps''' from '''Group_analysis/Scrambled-MEG''' to the Process1 window under the previous source maps. * Finally, drag and drop '''16 source maps''' from '''Group_analysis/Unfamiliar-MEG''' at the end of previous files. * Make sure you have the same number of files for each condition. * '''Note: '''PLS processes the data in “subject in condition” format. Therefore, all files from the same condition must be placed in the process window at once and files from the second condition must be added to the end of first condition, and so on for the rest of the conditions.<<BR>><<BR>> {{attachment:select_files2.gif}} <<BR>> Run process '''Test > Partial Least Squares (PLS) – More than Two Condition'''. * Enter the number of conditions and the number of subjects (files) per condition. * The other options are the same as explained above.<<BR>><<BR>> {{attachment:pipeline2.gif}} * The process will take some time. The results are displayed in the same way as before; however, you may have more bootstrap ratio files for different latent variables depending on the number of conditions you have. In this example, we have 2 latent variables.<<BR>><<BR>> {{attachment:results_overview2.gif}} <<BR>> * Check the p-values first to detect the significant latent variables. Each latent variable will have a p-value that indicates whether the related effect (or contrast) is significant or not. In this example, only latent variable 1 (LV1) is significant.<<BR>><<BR>> {{attachment:p_value2.gif}} <<BR>> * Open the contrast file and focus on the contrasts (effects) related to the significant latent variables.<<BR>><<BR>> {{attachment:contrast2.gif}} <<BR>> * Keep the bootstrap ratio files for significant latent variables and delete the rest. |
| Line 95: | Line 134: |
| 1. Khaligh-Razavi SM, Bainbridge W, Pantazis D, Oliva A (2016)<<BR>>[[http://biorxiv.org/content/early/2016/04/22/049700.abstract|From what we perceive to what we remember: Characterizing representational dynamics of visual memorability]]. ''bioRxiv'', 049700. 1. Cichy RM, Pantazis D, Oliva A (2014)<<BR>>[[http://www.nature.com/neuro/journal/v17/n3/full/nn.3635.html|Resolving human object recognition in space and time]], Nature Neuroscience, 17:455–462. |
1. Efron B, Tibshirani R (1986). "Bootstrap methods for standard errors, confidence intervals and other measures of statistical accuracy." ''Stat. Sci. 1'', 54– 77. 1. Krishnan A, Williams LJ, McIntosh AR, Abdi H (2011). "Partial Least Squares (PLS) methods for neuroimaging: a tutorial and review." ''Neuroimage, 56(2)'', 455-475. 1. McIntosh AR, Bookstein F, Haxby J, Grady C (1996). "Spatial pattern analysis of functional brain images using partial least squares." ''Neuroimage 3'', 143–157. 1. McIntosh AR, Lobaugh NJ (2004). "Partial least squares analysis of neuroimaging data: applications and advances." ''Neuroimage 23'', S250–S263. 1. Mišić B, Dunkley BT, Sedge PA, Da Costa L, Fatima Z, Berman MG, ... & Pang EW (2016). "Post-traumatic stress constrains the dynamic repertoire of neural activity." ''The Journal of Neuroscience, 36(2)'', 419-431. |
| Line 98: | Line 140: |
| <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/Decoding)>> | ---- |
Partial Least Squares (PLS)
Authors: Golia Shafiei
This tutorial explains the concept of Partial Least Squares (PLS) analysis in general, which was first introduced to the neuroimaging community in 1996 (McIntosh et al., 1996). In addition, we illustrate how to use PLS process on a sample data in Brainstorm.
Contents
License
PLS is a free toolbox that is a part of Baycrest, University of Toronto. The PLS code is written entirely in MATLAB (Mathworks Inc) and can be downloaded from https://www.rotman-baycrest.on.ca/index.php?section=345. To cite PLS Toolbox, please see the “References” section of this tutorial.
Introduction
Partial Least Squares (PLS) analysis is a multivariate statistical technique that is used to find the relationship between two blocks of variables. PLS that has various applications and types (Krishnan et al., 2011); however, the focus of this tutorial is on Mean-Centered PLS analysis, which is a common type of PLS while working with neuroimaging data. In this type of PLS analysis, one data block is neural activity (e.g. MEG measurements/source data here) while the other one is the experiment design (e.g. different groups/conditions).
PLS analysis is based on extracting the common information between the two data blocks by finding a correlation matrix and linear combinations of variables in both data blocks that have maximum covariance with one another. In the example provided here, we find a contrast between different conditions as well as patterns of brain activity that maximally covary with that specific contrast.
For this purpose, we take the neural activity as one data block, matrix X, where the rows of matrix X are observations (participants/trials) nested in conditions or groups, and the columns of X are variables that are arranged in a way that time scales are nested within sources. The other data block, matrix Y, is a matrix of dummy coding that is related to experimental design (different groups or conditions) (Krishnan et al., 2011).
PLS analysis first calculates a mean-centered matrix using matrices X and Y. Then, singular value decomposition (SVD) is applied on the mean-centered matrix. The outcome of PLS analysis is a set of latent variables that are in fact linear combinations of initial variables of the two data blocks that maximally covary with the resulting contrasts (Krishnan et al., 2011, Mišić et al., 2016).
Finally, the statistical significance of a latent variable is defined by a p-value calculated from permutation test. In addition, bootstrapping is used to assess the reliability of each original variable (e.g. a source at a time point) that contributes to the latent variable. Bootstrap ratios are calculated for each original variable for this purpose. More specifically, each latent variable consists of a set of singular values that describe the effect size, as well as a set of singular vectors, or weights, that define the contribution of each initial variable to the latent variables. The ratio of these weights to the standard errors estimated from bootstrapping is called bootstrap ratio. Therefore, the larger the magnitude of a bootstrap ratio, the larger the weight (i.e. contribution to the latent variable) and the smaller the standard error (i.e. higher stability) (McIntosh and Lobaugh, 2004, Mišić et al., 2016). Bootstrap ratio can be equivalent to a z-score if we have an approximately normal bootstrap distribution (Efron and Tibshirani, 1986).
PLS analysis was explained in general in this section. However, this tutorial assumes that the users are already familiar with basics of PLS analysis. If PLS is new to you or if you want to read more about PLS and its applications in details, please refer to the articles introduced in “References” section.
Download and installation
In order to run PLS process in Brainstorm, the PLS Toolbox must be downloaded from here and added to MATLAB pathway.
Data, Pre-Processing and Source Analysis
The data processed here is the same dataset that is used in MEG visual tutorial: Single subject and MEG visual tutorial: Group analysis. This dataset consists in simultaneous MEG/EEG recordings of 19 subjects performing a simple visual task on a large number of famous, unfamiliar and scrambled faces. The detailed presentation of experiment is available in the MEG visual tutorial: Single Subject.
You can follow this tutorial after processing the data as illustrated in MEG visual tutorial: Single Subject. Then:
Find the subject averages as it is explained in MEG visual tutorial: Group analysis by following the steps in the visual group analysis up to Section 7: Subject averages: Filter and normalize.
After you found all the averages across subjects, continue with Section 7 and filter the signals below 32Hz and extract time as it is explained. However, when filtering the sources, do not normalize the source values with respect to baseline (i.e. do not find z-score).
At this point, continue with the rest of the group analysis tutorial: Project sources on template and apply spatial smoothing on the source maps.
- Data is now ready for PLS analysis.
Running PLS
There are two PLS processes available in Brainstorm: * You can run PLS analysis for only two conditions through the Process2 tab at the bottom of the Brainstorm window.
You can also run PLS analysis for more than two conditions through the Process1 tab at the bottom of the Brainstorm window. This option is explained in the “Advanced” section of this tutorial.
Both of these two processes work in a similar way:
Input: the input files are the source data from different conditions. Number of samples per condition must be the same for all conditions. Make sure you have enough observations/trials per condition in order to get meaningful results from statistical tests.
Note: You can also run PLS analysis on sensor level data (channel data). However, the results will not be as meaningful and useful as the ones from source level data.
Output: the output files include bootstrap ratios and p-values for all latent variables. In addition, you can look at the contrast between conditions (groups and/or experimental tasks) for each latent variable. The output files are explained in details in the following sections.
We will continue with explaining the PLS process for only two conditions from now and we will leave PLS process for more conditions for the Advanced section of this tutorial.
Select files
- Select the process2 tab at the bottom of Brainstorm window.
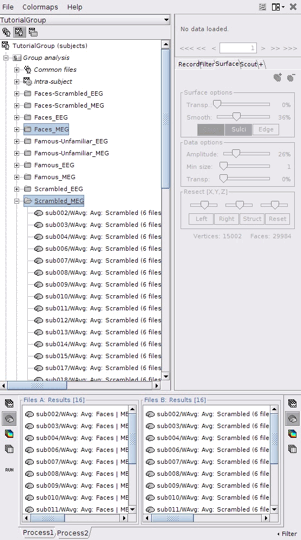
Drag and drop 16 source files from Group_analysis/Faces-MEG to the left (Files A).
Drag and drop 16 source files from Group_analysis/Scrambled-MEG to the right (Files B).
Note that the number of files in each window (“A” and “B”) must be the same.
Run PLS
- In Process2, click on [Run].
Select process Test > Partial Lest Squares (PLS). This opens the Pipeline editor window with the PLS process for two conditions.
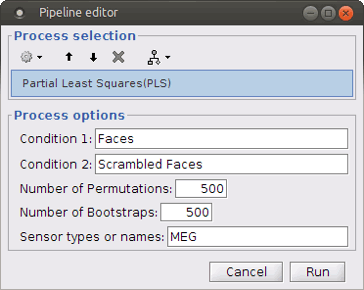
“Condition 1” and “Condition 2”: Name of the two input conditions: “Condition 1” and “Condition 2” are related to files in windows “A” and “B” of “Process2” tab, respectively. We can use “Faces” and “Scrambled Faces” as the condition names here.
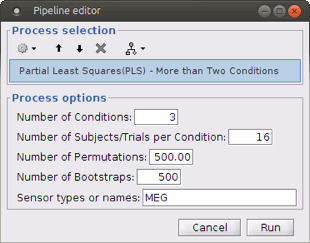
Number of Permutations: Indicates the number of permutations you want to run in PLS.
Number of Bootstraps: Indicates the number of bootstraps you want to run in PLS.
Sensor types or name: Indicates the type of sensors that has been used for source localization (e.g. MEG or EEG). This will occur in the result file name as well.
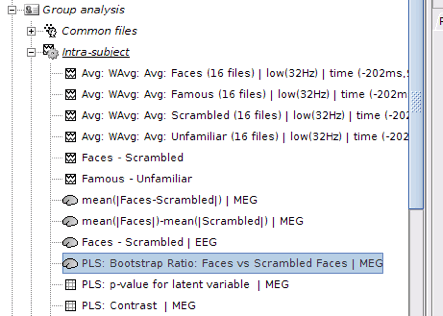
Run the process when the options are set. The process will take some time depending on the number of files, number of permutations and bootstraps. The results are then saved in Group Analysis > Intra-Subject folder.
PLS Results
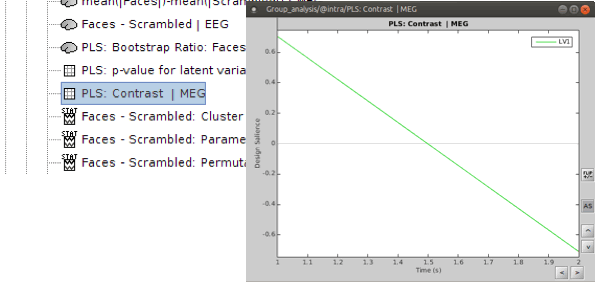
Output files include contrast between conditions, p-value of the latent variable and bootstrap ratios:
PLS : Contrast: You can first look at the contrast between two conditions: Double-click on the contrast file. The contrast is shown in the form of time series; however, the x-axis is not actual time. In fact, the integer numbers show the number of conditions (e.g. 1 for condition 1 or “Faces” and 2 for Condition 2 or “Scrambled Faces”) and the non-integer ones should be ignored.
PLS: p-value for latent variable: if you double click on this files, you will see a table containing the p-value of the latent variable that is related to the contrast shown above. A significant latent variable means the contrast observed between two conditions is also significant.
Note: You will see two numbers in the table for p-value of the latent variable when you run PLS for two conditions. Ignore the second one.
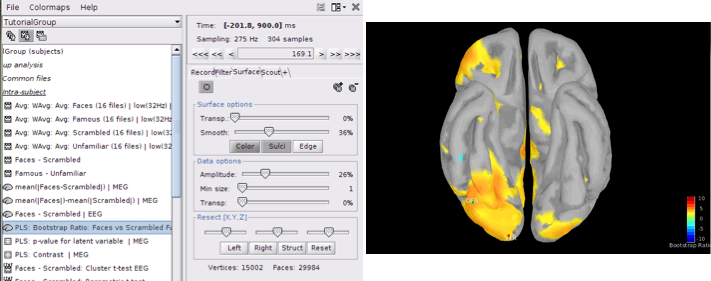
PLS: Bootstrap ratio: This file is saved in the format of source maps. Double-click on open the file. The colormap shows bootstrap ratios for each source at each time point. There are some important points you should keep in mind when working with bootstrap ratios:
Bootstrap ratios show how reliably each source is contributing to the observed contrast and latent variable at each time point. A bootstrap ratio larger than 2.58 is considered reliable. Therefore, adjust the amplitude of bootstrap ratios to 25%-26% to be able to see the reliable signals. For this, choose “Surface” tab from Brainstorm window and adjust the “Amplitude”.
- Do not display absolute values of the signals. Make sure “Absolute Values” is not selected in the colormap menu for this figure. The sign of bootstrap ratio is important since positive bootstrap ratios express the contrast as is, while bootstrap ratios with negative values express the opposite contrast.
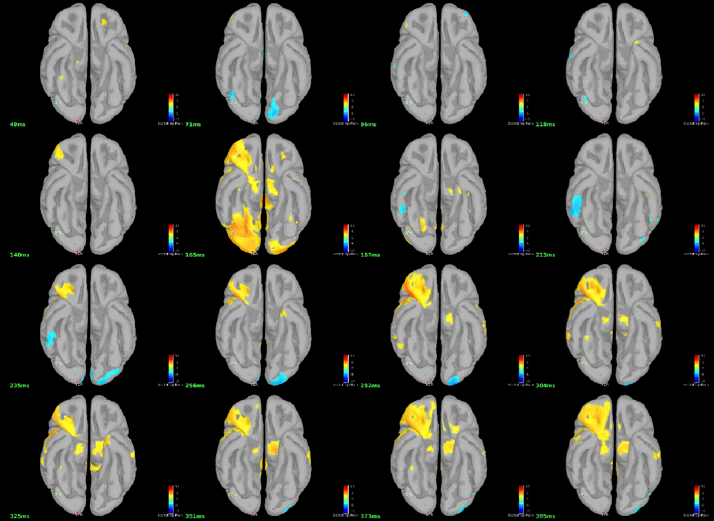
You can display the results at different time points using contact sheet.
You can also define regions of interest as usual: OFA (Occipital Face Area), V1.

Advanced Option
You can run PLS for more than 2 conditions through Process1 tab. However, you should be careful when arranging the files in this option. We will use the same dataset to illustrate how to use this option in Brainstorm.
- Select the Process1 tab at the bottom of Brainstorm window.
Drag and drop 16 source maps from Group_analysis/Famous-MEG to the Process1 window.
Drag and drop 16 source maps from Group_analysis/Scrambled-MEG to the Process1 window under the previous source maps.
Finally, drag and drop 16 source maps from Group_analysis/Unfamiliar-MEG at the end of previous files.
- Make sure you have the same number of files for each condition.
Note: PLS processes the data in “subject in condition” format. Therefore, all files from the same condition must be placed in the process window at once and files from the second condition must be added to the end of first condition, and so on for the rest of the conditions.

Run process Test > Partial Least Squares (PLS) – More than Two Condition.
- Enter the number of conditions and the number of subjects (files) per condition.
The other options are the same as explained above.
The process will take some time. The results are displayed in the same way as before; however, you may have more bootstrap ratio files for different latent variables depending on the number of conditions you have. In this example, we have 2 latent variables.
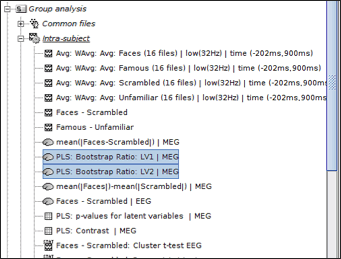
Check the p-values first to detect the significant latent variables. Each latent variable will have a p-value that indicates whether the related effect (or contrast) is significant or not. In this example, only latent variable 1 (LV1) is significant.

Open the contrast file and focus on the contrasts (effects) related to the significant latent variables.
- Keep the bootstrap ratio files for significant latent variables and delete the rest.
References
Efron B, Tibshirani R (1986). "Bootstrap methods for standard errors, confidence intervals and other measures of statistical accuracy." Stat. Sci. 1, 54– 77.
Krishnan A, Williams LJ, McIntosh AR, Abdi H (2011). "Partial Least Squares (PLS) methods for neuroimaging: a tutorial and review." Neuroimage, 56(2), 455-475.
McIntosh AR, Bookstein F, Haxby J, Grady C (1996). "Spatial pattern analysis of functional brain images using partial least squares." Neuroimage 3, 143–157.
McIntosh AR, Lobaugh NJ (2004). "Partial least squares analysis of neuroimaging data: applications and advances." Neuroimage 23, S250–S263.
Mišić B, Dunkley BT, Sedge PA, Da Costa L, Fatima Z, Berman MG, ... & Pang EW (2016). "Post-traumatic stress constrains the dynamic repertoire of neural activity." The Journal of Neuroscience, 36(2), 419-431.
